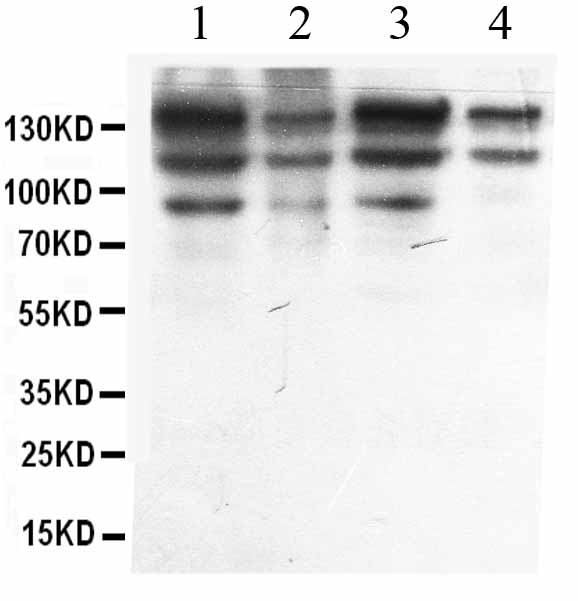
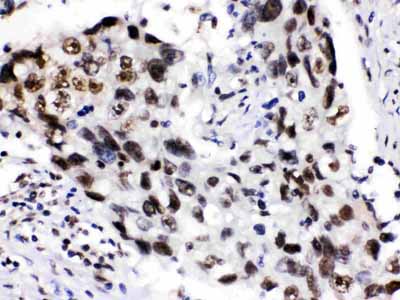
Anti-ADAR1 Picoband Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P |
|---|---|
| Primary Accession | P55265 |
| Host | Rabbit |
| Reactivity | Human |
| Clonality | Polyclonal |
| Format | Lyophilized |
| Description | Rabbit IgG polyclonal antibody for Double-stranded RNA-specific adenosine deaminase(ADAR) detection. Tested with WB, IHC-P in Human. |
| Reconstitution | Add 0.2ml of distilled water will yield a concentration of 500ug/ml. |
| Gene ID | 103 |
|---|---|
| Other Names | Double-stranded RNA-specific adenosine deaminase, DRADA, 3.5.4.37, 136 kDa double-stranded RNA-binding protein, p136, Interferon-inducible protein 4, IFI-4, K88DSRBP, ADAR, ADAR1, DSRAD, G1P1, IFI4 |
| Calculated MW | 136066 MW KDa |
| Application Details | Immunohistochemistry(Paraffin-embedded Section), 0.5-1 µg/ml, Human, By Heat Western blot, 0.1-0.5 µg/ml, Human |
| Subcellular Localization | Isoform 1: Cytoplasm. Nucleus. Shuttles between the cytoplasm and nucleus. |
| Tissue Specificity | Ubiquitously expressed, highest levels were found in brain and lung. Isoform 5 is expressed at higher levels in astrocytomas as compared to normal brain tissue and expression increases strikingly with the severity of the tumor, being higher in the most aggressive tumors. . |
| Protein Name | Double-stranded RNA-specific adenosine deaminase |
| Contents | Each vial contains 5mg BSA, 0.9mg NaCl, 0.2mg Na2HPO4, 0.05mg NaN3. |
| Immunogen | E.coli-derived human ADAR1 recombinant protein (Position: S128-Q346). Human ADAR1 shares 90.2% and 50.7% amino acid (aa) sequence identity with mouse and rat ADAR1, respectively. |
| Purification | Immunogen affinity purified. |
| Cross Reactivity | No cross reactivity with other proteins |
| Storage | At -20˚C for one year. After r˚Constitution, at 4˚C for one month. It˚Can also be aliquotted and stored frozen at -20˚C for a longer time.Avoid repeated freezing and thawing. |
| Name | ADAR |
|---|---|
| Synonyms | ADAR1, DSRAD, G1P1, IFI4 |
| Function | Catalyzes the hydrolytic deamination of adenosine to inosine in double-stranded RNA (dsRNA) referred to as A-to-I RNA editing (PubMed:12618436, PubMed:7565688, PubMed:7972084). This may affect gene expression and function in a number of ways that include mRNA translation by changing codons and hence the amino acid sequence of proteins since the translational machinery read the inosine as a guanosine; pre-mRNA splicing by altering splice site recognition sequences; RNA stability by changing sequences involved in nuclease recognition; genetic stability in the case of RNA virus genomes by changing sequences during viral RNA replication; and RNA structure- dependent activities such as microRNA production or targeting or protein-RNA interactions. Can edit both viral and cellular RNAs and can edit RNAs at multiple sites (hyper-editing) or at specific sites (site- specific editing). Its cellular RNA substrates include: bladder cancer- associated protein (BLCAP), neurotransmitter receptors for glutamate (GRIA2) and serotonin (HTR2C) and GABA receptor (GABRA3). Site-specific RNA editing of transcripts encoding these proteins results in amino acid substitutions which consequently alters their functional activities. Exhibits low-level editing at the GRIA2 Q/R site, but edits efficiently at the R/G site and HOTSPOT1. Its viral RNA substrates include: hepatitis C virus (HCV), vesicular stomatitis virus (VSV), measles virus (MV), hepatitis delta virus (HDV), and human immunodeficiency virus type 1 (HIV-1). Exhibits either a proviral (HDV, MV, VSV and HIV-1) or an antiviral effect (HCV) and this can be editing-dependent (HDV and HCV), editing-independent (VSV and MV) or both (HIV-1). Impairs HCV replication via RNA editing at multiple sites. Enhances the replication of MV, VSV and HIV-1 through an editing-independent mechanism via suppression of EIF2AK2/PKR activation and function. Stimulates both the release and infectivity of HIV-1 viral particles by an editing-dependent mechanism where it associates with viral RNAs and edits adenosines in the 5'UTR and the Rev and Tat coding sequence. Can enhance viral replication of HDV via A-to-I editing at a site designated as amber/W, thereby changing an UAG amber stop codon to an UIG tryptophan (W) codon that permits synthesis of the large delta antigen (L-HDAg) which has a key role in the assembly of viral particles. However, high levels of ADAR1 inhibit HDV replication. |
| Cellular Location | [Isoform 1]: Cytoplasm. Nucleus. Note=Shuttles between the cytoplasm and nucleus (PubMed:24753571, PubMed:7565688). Nuclear import is mediated by TNPO1 (PubMed:24753571). |
| Tissue Location | Ubiquitously expressed, highest levels were found in brain and lung (PubMed:7972084). Isoform 5 is expressed at higher levels in astrocytomas as compared to normal brain tissue and expression increases strikingly with the severity of the tumor, being higher in the most aggressive tumors. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Double-stranded RNA-specific adenosine deaminase, also known as ADAR1, is an enzyme that in humans is encoded by the ADAR gene. It is mapped to 1q21.3. This gene encodes the enzyme responsible for RNA editing by site-specific deamination of adenosines. This enzyme destabilizes double-stranded RNA through conversion of adenosine to inosine. Mutations in this gene have been associated with dyschromatosis symmetrica hereditaria. Alternative splicing results in multiple transcript variants.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.