Anti-CBFb Rabbit Monoclonal Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC, IF, ICC, FC |
|---|---|
| Primary Accession | Q13951 |
| Host | Rabbit |
| Isotype | IgG |
| Reactivity | Rat, Human, Mouse |
| Clonality | Monoclonal |
| Format | Liquid |
| Description | Anti-CBFb Rabbit Monoclonal Antibody . Tested in WB, IHC, ICC/IF, Flow Cytometry applications. This antibody reacts with Human, Mouse, Rat. |
| Gene ID | 865 |
|---|---|
| Other Names | Core-binding factor subunit beta, CBF-beta, Polyomavirus enhancer-binding protein 2 beta subunit, PEA2-beta, PEBP2-beta, SL3-3 enhancer factor 1 subunit beta, SL3/AKV core-binding factor beta subunit, CBFB |
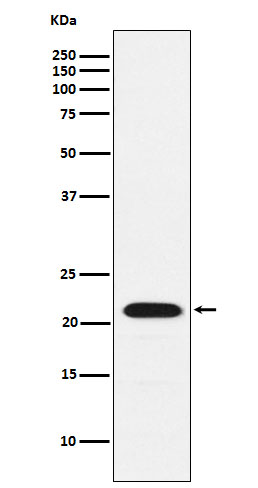
| Calculated MW | 22 kDa |
| Application Details | WB 1:500-1:1000 IHC 1:50-1:200 ICC/IF 1:50-1:200 FC 1:500 |
| Contents | Rabbit IgG in phosphate buffered saline, pH 7.4, 150mM NaCl, 0.02% sodium azide and 50% glycerol, 0.4-0.5mg/ml BSA. |
| Clone Names | Clone: 20C03 |
| Immunogen | A synthesized peptide derived from human CBFb |
| Purification | Affinity-chromatography |
| Storage | Store at -20°C for one year. For short term storage and frequent use, store at 4°C for up to one month. Avoid repeated freeze-thaw cycles. |
| Name | CBFB |
|---|---|
| Function | Forms the heterodimeric complex core-binding factor (CBF) with RUNX family proteins (RUNX1, RUNX2, and RUNX3). RUNX members modulate the transcription of their target genes through recognizing the core consensus binding sequence 5'-TGTGGT-3', or very rarely, 5'- TGCGGT-3', within their regulatory regions via their runt domain, while CBFB is a non-DNA-binding regulatory subunit that allosterically enhances the sequence-specific DNA-binding capacity of RUNX. The heterodimers bind to the core site of a number of enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T- cell receptor enhancers, LCK, IL3 and GM-CSF promoters. CBF complexes repress ZBTB7B transcription factor during cytotoxic (CD8+) T cell development. They bind to RUNX-binding sequence within the ZBTB7B locus acting as transcriptional silencer and allowing for cytotoxic T cell differentiation. |
| Cellular Location | Nucleus {ECO:0000250|UniProtKB:Q08024}. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.