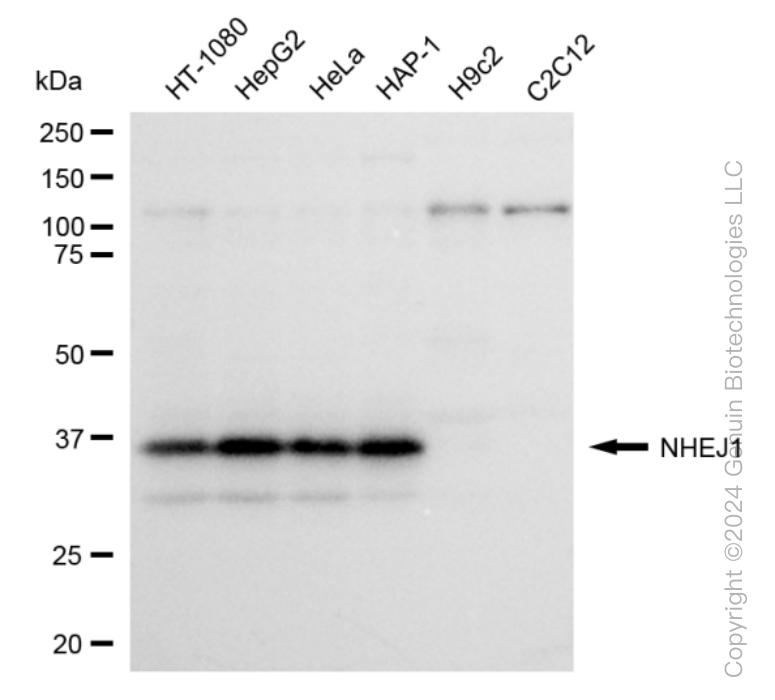
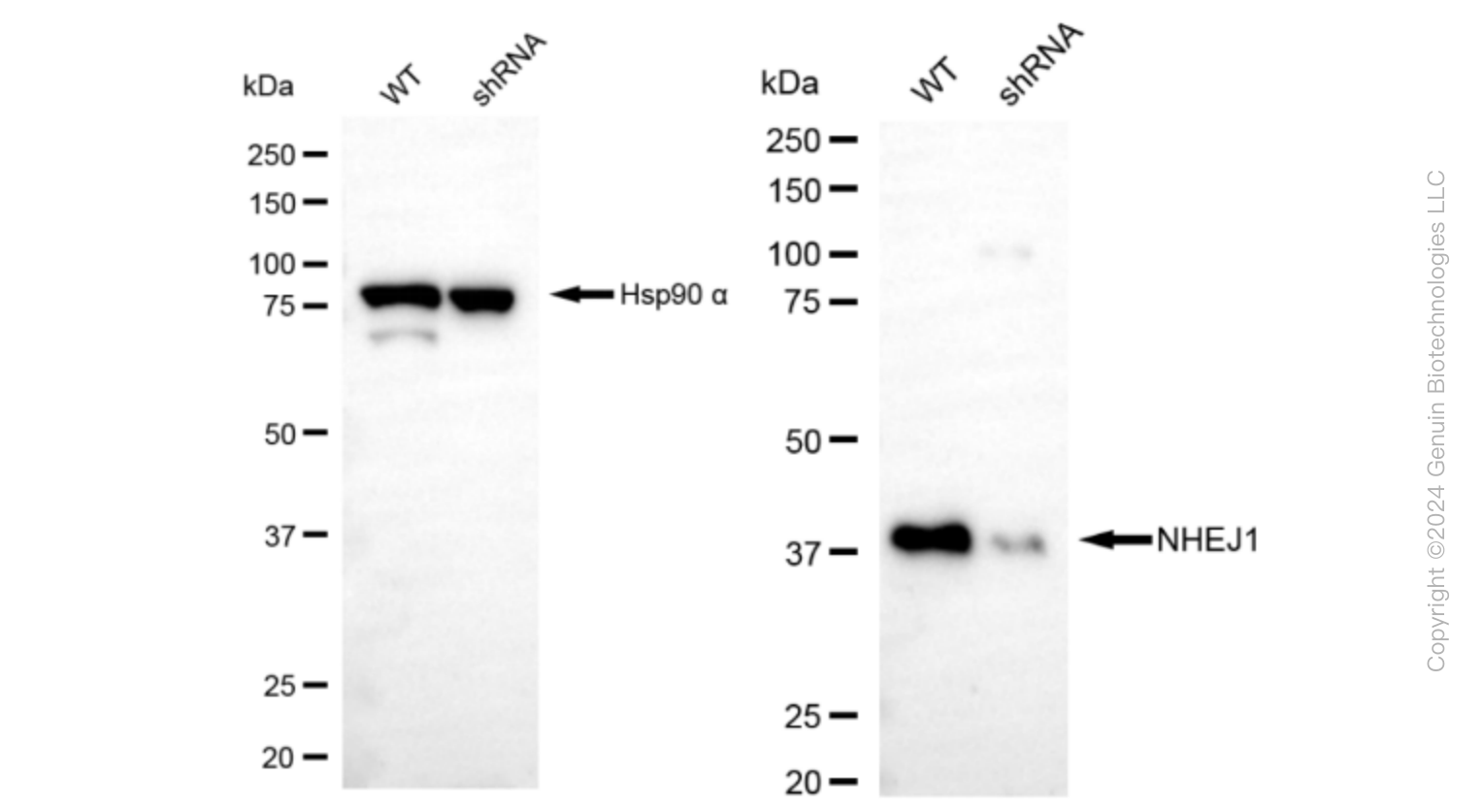
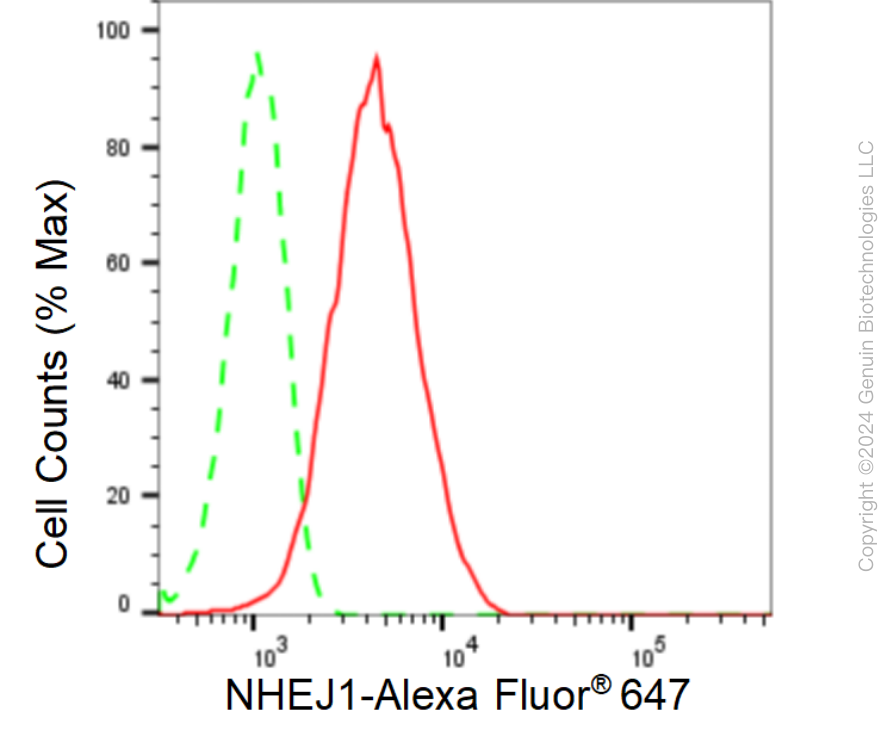
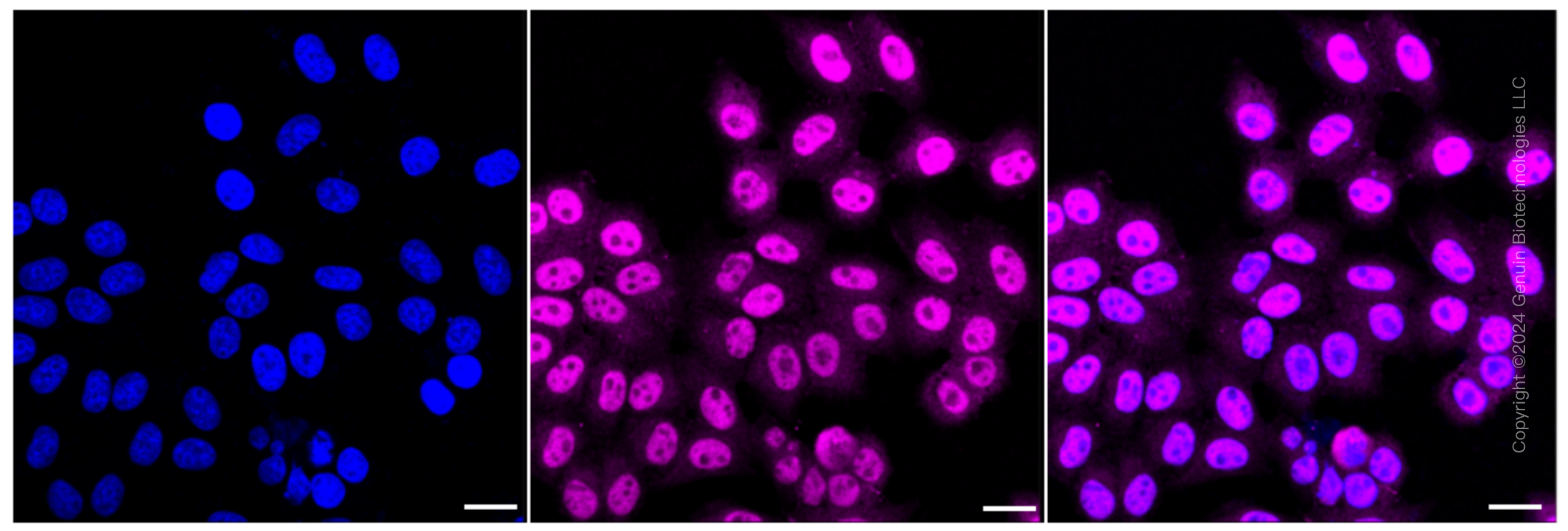
KD-Validated Anti-Non-Homologous End Joining Factor 1 Rabbit Monoclonal Antibody
Rabbit monoclonal antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, FC, ICC |
|---|---|
| Primary Accession | Q9H9Q4 |
| Reactivity | Human |
| Clonality | Monoclonal |
| Isotype | Rabbit IgG |
| Clone Names | 24GB480 |
| Calculated MW | Predicted, 33 kDa , observed , 39 kDa |
| Gene Name | NHEJ1 |
| Aliases | Non-Homologous End Joining Factor 1; XLF; XRCC4-Like Factor; Non-Homologous End-Joining Factor 1; Nonhomologous End-Joining Factor 1; Protein Cernunnos; Cernunnos; FLJ12610 |
| Immunogen | A synthesized peptide derived from human XLF |
| Gene ID | 79840 |
|---|---|
| Other Names | Non-homologous end-joining factor 1, Protein cernunnos, XRCC4-like factor, NHEJ1 {ECO:0000303|PubMed:17191205, ECO:0000312|HGNC:HGNC:25737} |
| Name | NHEJ1 {ECO:0000303|PubMed:17191205, ECO:0000312|HGNC:HGNC:25737} |
|---|---|
| Function | DNA repair protein involved in DNA non-homologous end joining (NHEJ); it is required for double-strand break (DSB) repair and V(D)J recombination and is also involved in telomere maintenance (PubMed:16439204, PubMed:16439205, PubMed:17317666, PubMed:17470781, PubMed:17717001, PubMed:18158905, PubMed:18644470, PubMed:20558749, PubMed:26100018, PubMed:28369633). Plays a key role in NHEJ by promoting the ligation of various mismatched and non-cohesive ends (PubMed:17470781, PubMed:17717001, PubMed:19056826). Together with PAXX, collaborates with DNA polymerase lambda (POLL) to promote joining of non-cohesive DNA ends (PubMed:25670504, PubMed:30250067). May act in concert with XRCC5-XRCC6 (Ku) to stimulate XRCC4-mediated joining of blunt ends and several types of mismatched ends that are non- complementary or partially complementary (PubMed:16439204, PubMed:16439205, PubMed:17317666, PubMed:17470781). In some studies, has been shown to associate with XRCC4 to form alternating helical filaments that bridge DNA and act like a bandage, holding together the broken DNA until it is repaired (PubMed:21768349, PubMed:21775435, PubMed:22228831, PubMed:22287571, PubMed:26100018, PubMed:27437582, PubMed:28500754). Alternatively, it has also been shown that rather than forming filaments, a single NHEJ1 dimer interacts through both head domains with XRCC4 to promote the close alignment of DNA ends (By similarity). The XRCC4-NHEJ1/XLF subcomplex binds to the DNA fragments of a DSB in a highly diffusive manner and robustly bridges two independent DNA molecules, holding the broken DNA fragments in close proximity to one other (PubMed:27437582, PubMed:28500754). The mobility of the bridges ensures that the ends remain accessible for further processing by other repair factors (PubMed:27437582). Binds DNA in a length-dependent manner (PubMed:17317666, PubMed:18158905). |
| Cellular Location | Nucleus. Chromosome. Note=Localizes to site of double-strand breaks; recruitment is dependent on XRCC5-XRCC6 (Ku) heterodimer |
| Tissue Location | Ubiquitously expressed. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.














 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.