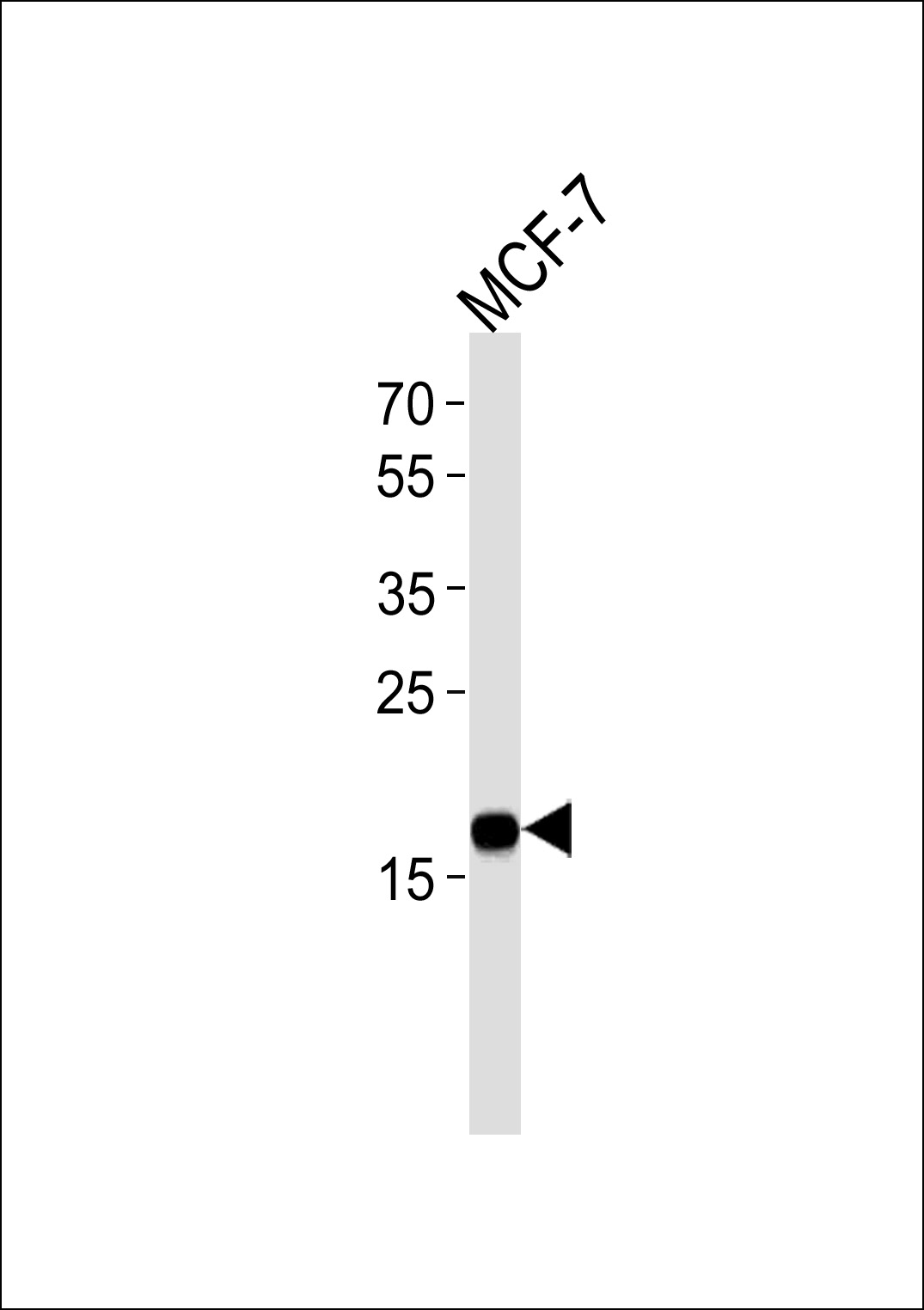
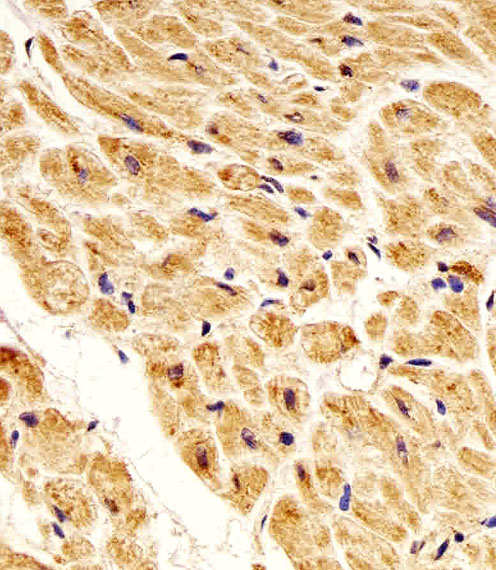
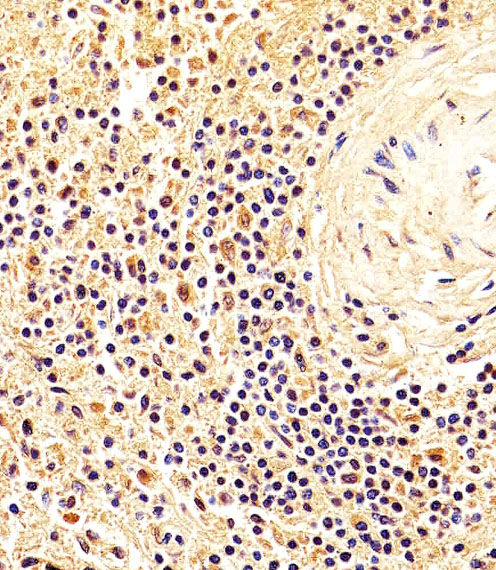
NME1 Antibody
Mouse Monoclonal Antibody (Mab)
- SPECIFICATION
- CITATIONS: 1
- PROTOCOLS
- BACKGROUND

Application
| IHC-P, WB, E |
|---|---|
| Primary Accession | P15531 |
| Reactivity | Human |
| Host | Mouse |
| Clonality | Monoclonal |
| Isotype | IgG2a |
| Clone/Animal Names | 1172CT2.4.1.1 |
| Calculated MW | 17149 Da |
| Gene ID | 4830 |
|---|---|
| Other Names | Nucleoside diphosphate kinase A, NDK A, NDP kinase A, Granzyme A-activated DNase, GAAD, Metastasis inhibition factor nm23, NM23-H1, Tumor metastatic process-associated protein, NME1, NDPKA, NM23 |
| Target/Specificity | Purified His-tagged NME1 protein was used to produced this monoclonal antibody. |
| Dilution | IHC-P~~1:25 WB~~1:1000 E~~Use at an assay dependent concentration. |
| Format | Purified monoclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein G column, followed by dialysis against PBS. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | NME1 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | NME1 (HGNC:7849) |
|---|---|
| Function | Catalyzes the transfer of a gamma-phosphoryl group from a nucleoside triphosphate, mainly ATP, to a nucleoside diphosphate via a ping-pong mechanism involving a phosphohistidine intermediate, therefore contributing to the nucleoside triphosphate homeostasis (PubMed:10952986, PubMed:14960567, PubMed:16313181, PubMed:1851158, PubMed:23519676, PubMed:33903070, PubMed:8810265, PubMed:9038158). Also phosphorylates geranyl pyrophosphate (GPP) and farnesyl pyrophosphate (FPP), linking it to isoprenoid metabolism (PubMed:10952986). Additionally, functions as a non-specific serine/threonine kinase and histidine protein kinase, transferring phosphoryl groups from its active site to target proteins (PubMed:8529641, PubMed:9038158). May function as a Mg(2+)-dependent single-stranded DNA endonuclease as part of the SET complex, cooperating with the 3'-5' exonuclease TREX1 to mediate apoptotic DNA fragmentation in cytotoxic T lymphocytes (PubMed:12628186, PubMed:16818237). Reported to nick one DNA strand, enabling TREX1 to remove nucleotides from the free 3' end, enhancing DNA damage and suppressing DNA end reannealing and repair (PubMed:16818237). Has been shown to cleave double strands DNA within the 3'-portions of both 5'-SHS silencer and NHE basal promoter element of the PDGFA gene, potentially repressing its transcription (PubMed:11694515). May also function as a Mg(2+)-dependent 3'-5' DNA exonuclease, excising nucleotides from 3' single-stranded DNA or DNA with 3' single strand overhangs, suggesting a role in DNA nucleolytic processing (PubMed:14960567, PubMed:16313181). Involved in the regulation of tumor metastasis and cellular differentiation (By similarity). Also required for cell motility (PubMed:8270257, PubMed:25582197). May control, with NME2, AcCoA usage between histone acetylation and fatty acid synthesis, possibly by binding and releasing AcCoA at transcriptionally active chromatin regions in proximity to histone acetyltransferase (HAT) (By similarity). |
| Cellular Location | Cytoplasm. Nucleus. Cell membrane {ECO:0000250|UniProtKB:P52175}. Note=Cell-cycle dependent nuclear localization which can be induced by degradation of the SET complex by GzmA (PubMed:12628186). In response to DNA damage, translocates to the nucleus where it might participate in DNA nucleolytic processing (PubMed:16313181). |
| Tissue Location | Ubiquitously expressed (PubMed:12601555, PubMed:16442775). Expressed in tumor cell lines (PubMed:10512675, PubMed:16442775). |

Provided below are standard protocols that you may find useful for product applications.
Background
Major role in the synthesis of nucleoside triphosphates other than ATP. Possesses nucleoside-diphosphate kinase, serine/threonine-specific protein kinase, geranyl and farnesyl pyrophosphate kinase, histidine protein kinase and 3'-5' exonuclease activities. Involved in cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor endocytosis, and gene expression. Required for neural development including neural patterning and cell fate determination.
References
Rosengard A.M., et al. Nature 342:177-180(1989).
Gilles A.-M., et al. J. Biol. Chem. 266:8784-8789(1991).
Wang L., et al. Cancer Res. 53:717-720(1993).
Dooley S., et al. Hum. Genet. 93:63-66(1994).
Ni X., et al. J. Hum. Genet. 48:96-100(2003).
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.














 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.