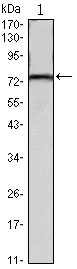
KLHL22 Antibody
Purified Mouse Monoclonal Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, ICC, E |
|---|---|
| Primary Accession | Q53GT1 |
| Reactivity | Human |
| Host | Mouse |
| Clonality | Monoclonal |
| Clone Names | 7B10 |
| Isotype | IgG1 |
| Calculated MW | 72kDa |
| Description | KLHL22 (kelch-like protein 22) is a 634 amino acid protein that is related to the Drosophilakelch protein, which is required to maintain Actin organization in ovarian ring canals. Mutations affecting Kelch function result in the failure of Kelch to associate with the ring canals and subsequent female sterility. Human KLHL22 protein contains six kelch repeats and one BTB (POZ) domain. The BTB (Broad-Complex, Tramtrack and Bric a brac) domain, also known as the POZ (Poxvirus and Zinc finger) domain, is an N-terminal homodimerization domain that contains multiple copies of kelch repeats and/or C2H2-type zinc fingers. Proteins that contain BTB domains are thought to be involved in transcriptional regulation via control of chromatin structure and function. There are two isoforms of KLHL22 that are produced as a result of alternative splicing events. |
| Immunogen | Purified recombinant fragment of human KLHL22 expressed in E. Coli. |
| Formulation | Ascitic fluid containing 0.03% sodium azide. |
| Gene ID | 84861 |
|---|---|
| Other Names | Kelch-like protein 22, KLHL22 |
| Dilution | WB~~1/500 - 1/2000 ICC~~N/A E~~N/A |
| Storage | Maintain refrigerated at 2-8°C for up to 6 months. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | KLHL22 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | KLHL22 (HGNC:25888) |
|---|---|
| Function | Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin ligase complex required for chromosome alignment and localization of PLK1 at kinetochores. The BCR(KLHL22) ubiquitin ligase complex mediates monoubiquitination of PLK1, leading to PLK1 dissociation from phosphoreceptor proteins and subsequent removal from kinetochores, allowing silencing of the spindle assembly checkpoint (SAC) and chromosome segregation. Monoubiquitination of PLK1 does not lead to PLK1 degradation (PubMed:19995937, PubMed:23455478). The BCR(KLHL22) ubiquitin ligase complex is also responsible for the amino acid-stimulated 'Lys-48' polyubiquitination and proteasomal degradation of DEPDC5. Through the degradation of DEPDC5, releases the GATOR1 complex-mediated inhibition of the TORC1 pathway. It is therefore an amino acid-dependent activator within the amino acid-sensing branch of the TORC1 pathway, indirectly regulating different cellular processes including cell growth and autophagy (PubMed:29769719). |
| Cellular Location | Cytoplasm, cytosol. Cytoplasm, cytoskeleton, microtubule organizing center, centrosome. Cytoplasm, cytoskeleton, spindle. Nucleus. Lysosome Note=Mainly cytoplasmic in prophase and prometaphase. Associates with the mitotic spindle as the cells reach chromosome bi-orientation Localizes to the centrosomes shortly before cells enter anaphase After anaphase onset, predominantly associates with the polar microtubules connecting the 2 opposing centrosomes and gradually diffuses into the cytoplasm during telophase (PubMed:23455478). Localizes to the nucleus upon amino acid starvation (PubMed:29769719). Relocalizes to the cytosol and associates with lysosomes when amino acids are available (PubMed:29769719). |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
References
1. Gene. 1994 Jan 28;138(1-2):171-4. 2. Nat Genet. 2004 Jan;36(1):40-5. 3. Cell. 2009 Jul 23;138(2):389-403.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.