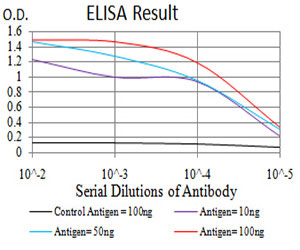
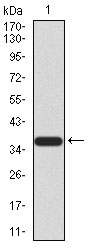
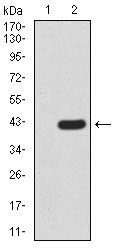
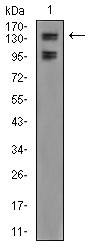
FBXL10 Antibody
Purified Mouse Monoclonal Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
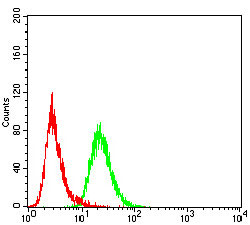
| WB, FC, E |
|---|---|
| Primary Accession | Q8NHM5 |
| Reactivity | Human |
| Host | Mouse |
| Clonality | Monoclonal |
| Clone Names | 6F6G11 |
| Isotype | IgG2b |
| Calculated MW | 152.6kDa |
| Description | This gene encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. The F-box proteins are divided into 3 classes: Fbws containing WD-40 domains, Fbls containing leucine-rich repeats, and Fbxs containing either different protein-protein interaction modules or no recognizable motifs. The protein encoded by this gene belongs to the Fbls class. Multiple alternatively spliced transcript variants have been found for this gene, but the full-length nature of some variants has not been determined. |
| Immunogen | Purified recombinant fragment of human FBXL10 (AA: 457-555) expressed in E. Coli. |
| Formulation | Purified antibody in PBS with 0.05% sodium azide |
| Gene ID | 84678 |
|---|---|
| Other Names | Lysine-specific demethylase 2B, 1.14.11.27, CXXC-type zinc finger protein 2, F-box and leucine-rich repeat protein 10, F-box protein FBL10, F-box/LRR-repeat protein 10, JmjC domain-containing histone demethylation protein 1B, Jumonji domain-containing EMSY-interactor methyltransferase motif protein, Protein JEMMA, Protein-containing CXXC domain 2, [Histone-H3]-lysine-36 demethylase 1B, KDM2B, CXXC2, FBL10, FBXL10, JHDM1B, PCCX2 |
| Dilution | WB~~1/500 - 1/2000 FC~~1/200 - 1/400 E~~1/10000 |
| Storage | Maintain refrigerated at 2-8°C for up to 6 months. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | FBXL10 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | KDM2B |
|---|---|
| Function | Histone demethylase that demethylates 'Lys-4' and 'Lys-36' of histone H3, thereby playing a central role in histone code (PubMed:16362057, PubMed:17994099, PubMed:26237645). Preferentially demethylates trimethylated H3 'Lys-4' and dimethylated H3 'Lys-36' residue while it has weak or no activity for mono- and tri-methylated H3 'Lys-36' (PubMed:16362057, PubMed:17994099, PubMed:26237645). Preferentially binds the transcribed region of ribosomal RNA and represses the transcription of ribosomal RNA genes which inhibits cell growth and proliferation (PubMed:16362057, PubMed:17994099). May also serve as a substrate-recognition component of the SCF (SKP1-CUL1-F-box protein)-type E3 ubiquitin ligase complex (Probable). |
| Cellular Location | Nucleus, nucleolus. Nucleus. Chromosome |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
References
1.Cancer Res. 2014 Jul 15;74(14):3935-46.2.Elife. 2012 Dec 18;1:e00205.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.