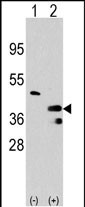
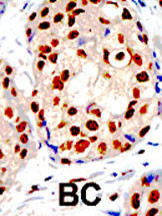
PRMT1 Antibody (C-term)
Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| IHC-P, WB, E |
|---|---|
| Primary Accession | Q99873 |
| Other Accession | Q63009, Q9JIF0 |
| Reactivity | Human |
| Predicted | Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | Rabbit IgG |
| Calculated MW | 42462 Da |
| Antigen Region | 326-357 aa |
| Gene ID | 3276 |
|---|---|
| Other Names | Protein arginine N-methyltransferase 1, 211-, Histone-arginine N-methyltransferase PRMT1, Interferon receptor 1-bound protein 4, PRMT1, HMT2, HRMT1L2, IR1B4 |
| Target/Specificity | This PRMT1 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 316-347 amino acids from the C-terminal region of human PRMT1. |
| Dilution | IHC-P~~1:50~100 WB~~1:1000 E~~Use at an assay dependent concentration. |
| Format | Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is prepared by Saturated Ammonium Sulfate (SAS) precipitation followed by dialysis against PBS. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | PRMT1 Antibody (C-term) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | PRMT1 (HGNC:5187) |
|---|---|
| Function | Arginine methyltransferase that methylates (mono and asymmetric dimethylation) the guanidino nitrogens of arginyl residues present in proteins such as ESR1, histone H2, H3 and H4, FMR1, ILF3, HNRNPA1, HNRNPD, NFATC2IP, SUPT5H, TAF15, EWS, HABP4, SERBP1, RBM15, FOXO1, CHTOP, MAP3K5/ASK1, MICU1 and NPRL2 (PubMed:10749851, PubMed:15741314, PubMed:16879614, PubMed:18951090, PubMed:22095282, PubMed:25284789, PubMed:26575292, PubMed:26876602, PubMed:27642082, PubMed:30765518, PubMed:31257072, PubMed:38006878). Constitutes the main enzyme that mediates monomethylation and asymmetric dimethylation of histone H4 'Arg-3' (H4R3me1 and H4R3me2a, respectively), a specific tag for epigenetic transcriptional activation. May be involved in the regulation of TAF15 transcriptional activity, act as an activator of estrogen receptor (ER)-mediated transactivation, play a key role in neurite outgrowth and act as a negative regulator of megakaryocytic differentiation, by modulating p38 MAPK pathway. Methylates RBM15, promoting ubiquitination and degradation of RBM15 (PubMed:26575292). Methylates MRE11 and TP53BP1, promoting the DNA damage response (PubMed:15741314, PubMed:16294045, PubMed:29651020). Methylates FOXO1 and retains it in the nucleus increasing its transcriptional activity (PubMed:18951090). Methylates CHTOP and this methylation is critical for its 5-hydroxymethylcytosine (5hmC)-binding activity (PubMed:25284789). Methylates MAP3K5/ASK1 at 'Arg-78' and 'Arg-80' which promotes association of MAP3K5 with thioredoxin and negatively regulates MAP3K5 association with TRAF2, inhibiting MAP3K5 stimulation and MAP3K5-induced activation of JNK (PubMed:22095282). Methylates H4R3 in genes involved in glioblastomagenesis in a CHTOP- and/or TET1- dependent manner (PubMed:25284789). Plays a role in regulating alternative splicing in the heart (By similarity). Methylates NPRL2 at 'Arg-78' leading to inhibition of its GTPase activator activity and then the GATOR1 complex and consequently inducing timely mTORC1 activation under methionine-sufficient conditions (PubMed:38006878). |
| Cellular Location | Nucleus. Nucleus, nucleoplasm {ECO:0000250|UniProtKB:Q9JIF0}. Cytoplasm. Cytoplasm, cytosol {ECO:0000250|UniProtKB:Q9JIF0}. Lysosome membrane. Note=Mostly found in the cytoplasm Colocalizes with CHTOP within the nucleus. Low levels detected also in the chromatin fraction (By similarity). Upon methionine stimulation, localizes to the lysosome membrane in an NPRL2-dependent manner (PubMed:38006878). {ECO:0000250|UniProtKB:Q9JIF0, ECO:0000269|PubMed:38006878} |
| Tissue Location | Widely expressed (PubMed:11097842). Expressed strongly in colorectal cancer cells (at protein level) (PubMed:28040436). Expressed strongly in colorectal cancer tissues compared to wild-type colon samples (at protein level) (PubMed:28040436). Expressed strongly in colorectal cancer tissues compared to wild-type colon samples (PubMed:28040436) |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Arginine methylation is an irreversible post translational modification which has only recently been linked to protein activity. At least three types of PRMT enzymes have been identified in mammalian cells. These enzymes have been shown to have essential regulatory functions by methylation of key proteins in several fundamental areas. These protein include nuclear proteins, IL enhancer binding factor, nuclear factors, cell cycle proteins, signal transduction proteins, apoptosis proteins, and viral proteins. The mammalian PRMT family currently consists of 7 members that share two large domains of homology. Outside of these domains, epitopes were identified and antibodies against all 7 PRMT members have been developed.
References
Zhang, X., et al., EMBO J. 19(14):3509-3519 (2000).
Scorilas, A., et al., Biochem. Biophys. Res. Commun. 278(2):349-359 (2000).
Scott, H.S., et al., Genomics 48(3):330-340 (1998).
Abramovich, C., et al., EMBO J. 16(2):260-266 (1997).
Nikawa, J., et al., Gene 171(1):107-111 (1996).
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.