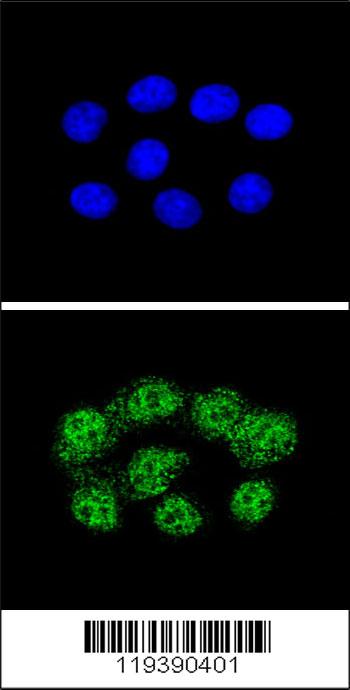
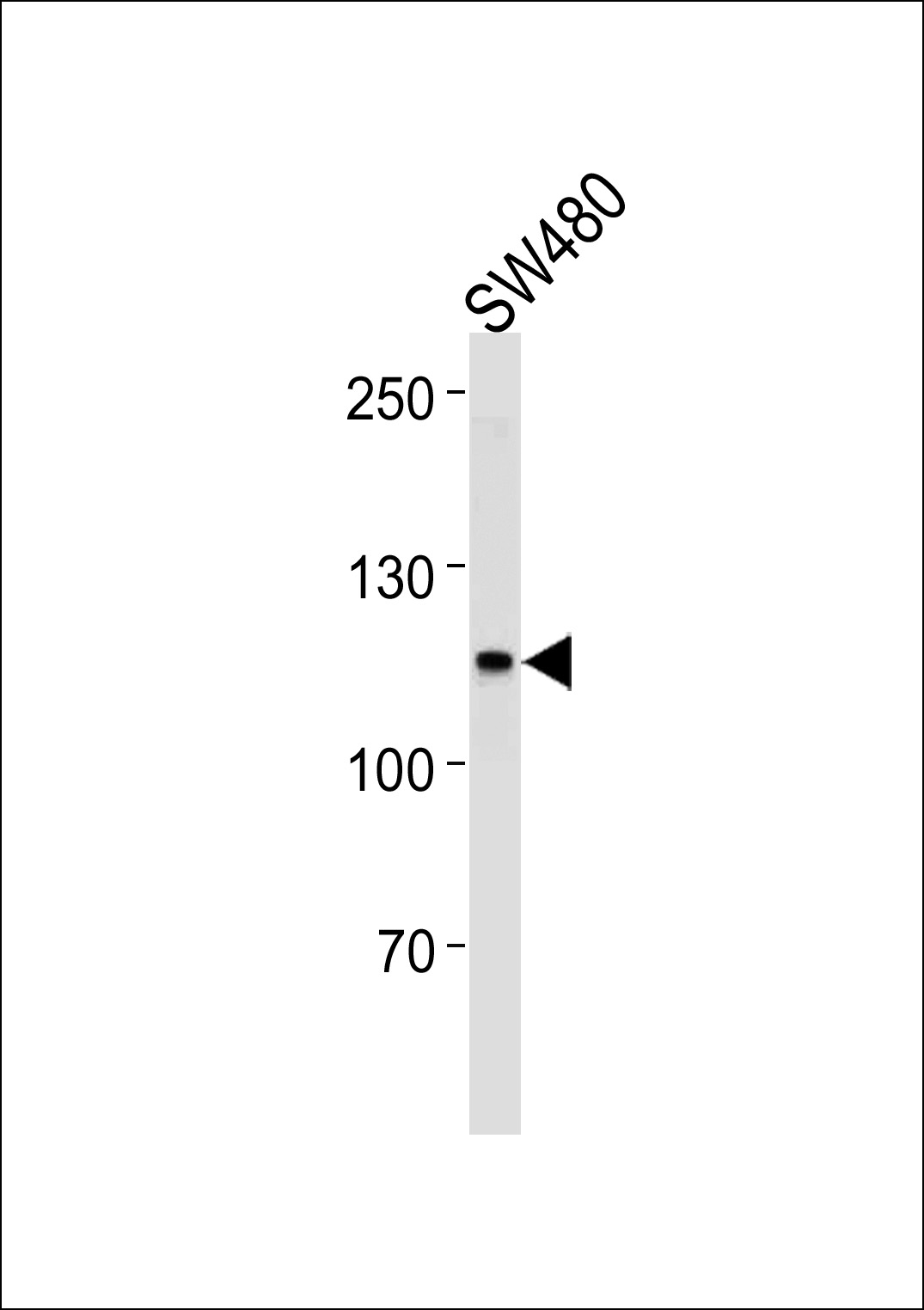
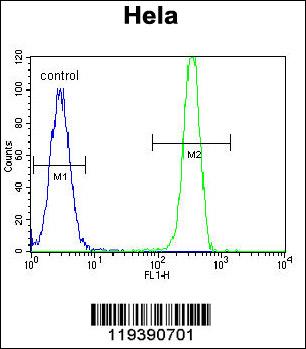
MSH2 Antibody (Center)
Affinity Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IF, FC, E |
|---|---|
| Primary Accession | P43246 |
| Other Accession | NP_000242.1 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | Rabbit IgG |
| Calculated MW | 104743 Da |
| Antigen Region | 637-665 aa |
| Gene ID | 4436 |
|---|---|
| Other Names | DNA mismatch repair protein Msh2, hMSH2, MutS protein homolog 2, MSH2 |
| Target/Specificity | This MSH2 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 637-665 amino acids from the Central region of human MSH2. |
| Dilution | WB~~1:1000 IF~~1:10~50 FC~~1:10~50 E~~Use at an assay dependent concentration. |
| Format | Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | MSH2 Antibody (Center) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | MSH2 |
|---|---|
| Function | Component of the post-replicative DNA mismatch repair system (MMR). Forms two different heterodimers: MutS alpha (MSH2-MSH6 heterodimer) and MutS beta (MSH2-MSH3 heterodimer) which binds to DNA mismatches thereby initiating DNA repair. When bound, heterodimers bend the DNA helix and shields approximately 20 base pairs. MutS alpha recognizes single base mismatches and dinucleotide insertion-deletion loops (IDL) in the DNA. MutS beta recognizes larger insertion-deletion loops up to 13 nucleotides long. After mismatch binding, MutS alpha or beta forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. Recruits DNA helicase MCM9 to chromatin which unwinds the mismatch containing DNA strand (PubMed:26300262). ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The ATPase activity associated with MutS alpha regulates binding similar to a molecular switch: mismatched DNA provokes ADP-->ATP exchange, resulting in a discernible conformational transition that converts MutS alpha into a sliding clamp capable of hydrolysis-independent diffusion along the DNA backbone. This transition is crucial for mismatch repair. MutS alpha may also play a role in DNA homologous recombination repair. In melanocytes may modulate both UV-B-induced cell cycle regulation and apoptosis. |
| Cellular Location | Nucleus. Chromosome |
| Tissue Location | Ubiquitously expressed. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
MSH2 was identified as a locus frequently mutated in hereditary nonpolyposis colon cancer (HNPCC). When cloned, it was discovered to be a human homolog of the E. coli mismatch repair gene mutS, consistent with the characteristic alterations in microsatellite sequences (RER+ phenotype) found in HNPCC. [provided by RefSeq].
References
Kim, M., et al. Cancer Sci. 101(11):2436-2442(2010)
Mangoni, M., et al. Int. J. Radiat. Oncol. Biol. Phys. (2010) In press :
Srivastava, K., et al. Cancer 116(13):3160-3169(2010)
van der Post, R.S., et al. J. Med. Genet. 47(7):464-470(2010)
Langner, E., et al. J. Genet. 89(1):101-104(2010)
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.