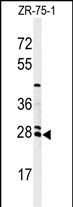
NAT8 Antibody (Center)
Affinity Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS: 2
- PROTOCOLS
- BACKGROUND

Application
| IHC-P, WB, E |
|---|---|
| Primary Accession | Q9UHE5 |
| Other Accession | Q9UHF3 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | Rabbit IgG |
| Calculated MW | 25619 Da |
| Antigen Region | 110-138 aa |
| Gene ID | 9027 |
|---|---|
| Other Names | N-acetyltransferase 8, 231-, Acetyltransferase 2, ATase2, Camello-like protein 1, Cysteinyl-conjugate N-acetyltransferase, CCNAT, NAT8 (HGNC:18069) |
| Target/Specificity | This NAT8 antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 110-138 amino acids from the Central region of human NAT8. |
| Dilution | IHC-P~~1:50~100 WB~~1:1000 E~~Use at an assay dependent concentration. |
| Format | Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | NAT8 Antibody (Center) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | NAT8 (HGNC:18069) |
|---|---|
| Function | Endoplasmic reticulum (ER)-membrane-bound lysine N- acetyltransferase catalyzing the N6-acetylation of lysine residues in the lumen of the ER in various proteins, including PROM1 and BACE1, using acetyl-CoA as acetyl donor (PubMed:19011241, PubMed:22267734, PubMed:24556617, PubMed:31945187). Thereby, may regulate apoptosis through the acetylation and the regulation of the expression of PROM1 (PubMed:24556617). May also regulate amyloid beta-peptide secretion through acetylation of BACE1 and the regulation of its expression in neurons (PubMed:19011241). N(6)-lysine acetylation in the ER maintains protein homeostasis and regulates reticulophagy (By similarity). Alternatively, acetylates the free alpha-amino group of cysteine S- conjugates to form mercapturic acids (PubMed:20392701). This is the final step in a major route for detoxification of a wide variety of reactive electrophiles which starts with their incorporation into glutathione S-conjugates. The glutathione S-conjugates are then further processed into cysteine S-conjugates and finally mercapturic acids which are water soluble and can be readily excreted in urine or bile. |
| Cellular Location | Endoplasmic reticulum-Golgi intermediate compartment membrane; Single-pass type II membrane protein. Endoplasmic reticulum membrane; Single-pass type II membrane protein |
| Tissue Location | Preferentially expressed in liver and kidney. Also detected in brain (at protein level). |

Provided below are standard protocols that you may find useful for product applications.
Background
This protein, isolated using the differential display method to detect tissue-specific genes, is specifically expressed in kidney and liver. The encoded protein shows amino acid sequence similarity to N-acetyltransferases. A similar protein in Xenopus affects cell adhesion and gastrulation movements, and may be localized in the secretory pathway. A highly similar paralog is found in a cluster with this gene.
References
Ko, M.H., et al. J. Biol. Chem. 284(4):2482-2492(2009) Juhanson, P., et al. BMC Med. Genet. 9, 25 (2008) Barrios-Rodiles, M., et al. Science 307(5715):1621-1625(2005)
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.














 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.