Rabbit Anti-ATF4/CREB-2 Polyclonal Antibody
Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
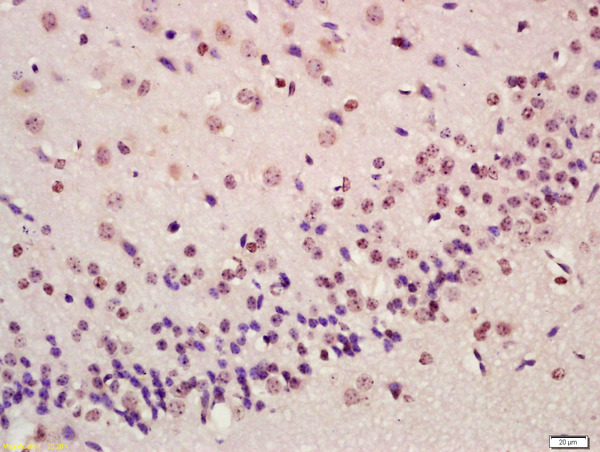
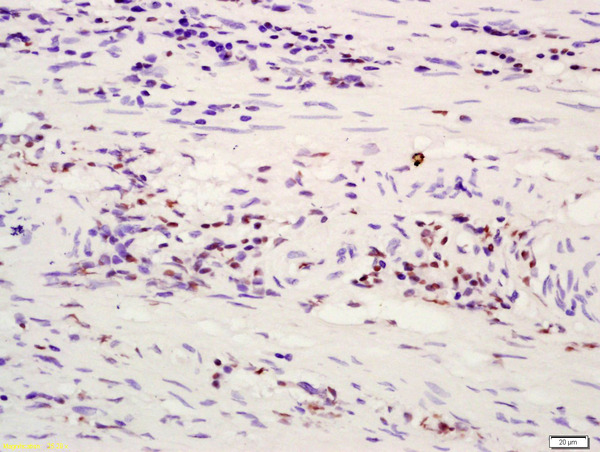
| WB, IHC-P, IHC-F, IF, ICC, E |
|---|---|
| Primary Accession | P18848 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
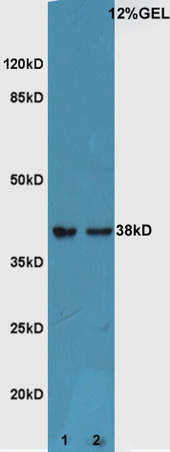
| Calculated MW | 38 KDa |
| Physical State | Liquid |
| Immunogen | KLH conjugated synthetic peptide derived from human ATF4 |
| Epitope Specificity | 251-351/351 |
| Isotype | IgG |
| Purity | affinity purified by Protein A |
| Buffer | 0.01M TBS (pH7.4) with 1% BSA, 0.02% Proclin300 and 50% Glycerol. |
| SUBCELLULAR LOCATION | Cytoplasm. Cell membrane. Nucleus. Colocalizes with GABBR1 in hippocampal neuron dendritic membranes. |
| SIMILARITY | Belongs to the bZIP family. Contains 1 bZIP domain. |
| SUBUNIT | Binds DNA as a homo- or heterodimer. Interacts (via its leucine zipper domain) with GABBR1 and GABBR2 (via their C-termini). Interacts (via its DNA binding domain) with FOXO1 (C-terminal half); the interaction occurs in osteoblasts and regulates glucose homeostasis through suppression of beta-cell proliferation and a decrease in insulin production. Interacts with SATB2; the interaction results in enhanced DNA binding and transactivation by these transcription factors. Interacts with CEP290 (via an N-terminal region). Interacts with NEK6, DAPK2 (isoform 2) and ZIPK/DAPK3. Forms a heterodimer with TXLNG in osteoblasts. Interacts with DDIT3/CHOP. |
| Post-translational modifications | Ubiquitinated by SCF(BTRC) in response to mTORC1 signal, followed by proteasomal degradation and leading to down-regulate expression of SIRT4. Phosphorylated by NEK6. Phosphorylated on the betaTrCP degron motif at Ser-219, followed by phosphorylation at Thr-213, Ser-224, Ser-231, Ser-235 and Ser-248, promoting interaction with BTRC and ubiquitination. Phosphorylation is promoted by mTORC1. Phosphorylated by NEK6. |
| Important Note | This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |
| Background Descriptions | ATF4 is a transcription factor that was originally identified as a widely expressed mammalian DNA binding protein that could bind a tax-responsive enhancer element in the LTR of HTLV1. The encoded protein was also isolated and characterized as the cAMP-response element binding protein 2 (CREB2). The protein encoded by this gene belongs to a family of DNA-binding proteins that includes the AP1 family of transcription factors, cAMP-response element binding proteins (CREBs) and CREB-like proteins. These transcription factors share a leucine zipper region that is involved in protein-protein interactions, located C-terminal to a stretch of basic amino acids that functions as a DNA binding domain (referenced from Entrez gene). |
| Gene ID | 468 |
|---|---|
| Other Names | CREB2; TXREB; CREB-2; TAXREB67; Cyclic AMP-dependent transcription factor ATF-4; cAMP-dependent transcription factor ATF-4; Activating transcription factor 4; Cyclic AMP-responsive element-binding protein 2; cAMP-responsive element-binding protein 2; DNA-binding protein TAXREB67; Tax-responsive enhancer element-binding protein 67; ATF4 |
| Dilution | WB=1:500-2000,IHC-P=1:100-500,IHC-F=1:100-500,ICC=1:100,IF=1:100-500,Flow-Cyt=0.2 µg /test,ELISA=1:5000-10000 |
| Format | 0.01M TBS(pH7.4), 0.09% (W/V) sodium azide and 50% Glyce |
| Storage | Store at -20 ℃ for one year. Avoid repeated freeze/thaw cycles. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 ℃. |
| Name | ATF4 {ECO:0000303|PubMed:2516827, ECO:0000312|HGNC:HGNC:786} |
|---|---|
| Function | Transcription factor that binds the cAMP response element (CRE) (consensus: 5'-GTGACGT[AC][AG]-3') and displays two biological functions, as regulator of metabolic and redox processes under normal cellular conditions, and as master transcription factor during integrated stress response (ISR) (PubMed:16682973, PubMed:17684156, PubMed:31023583, PubMed:31444471, PubMed:32132707). Binds to asymmetric CRE's as a heterodimer and to palindromic CRE's as a homodimer (By similarity). Core effector of the ISR, which is required for adaptation to various stress such as endoplasmic reticulum (ER) stress, amino acid starvation, mitochondrial stress or oxidative stress (PubMed:31023583, PubMed:32132707). During ISR, ATF4 translation is induced via an alternative ribosome translation re-initiation mechanism in response to EIF2S1/eIF-2-alpha phosphorylation, and stress-induced ATF4 acts as a master transcription factor of stress-responsive genes in order to promote cell recovery (PubMed:31023583, PubMed:32132706, PubMed:32132707). Promotes the transcription of genes linked to amino acid sufficiency and resistance to oxidative stress to protect cells against metabolic consequences of ER oxidation (By similarity). Activates the transcription of NLRP1, possibly in concert with other factors in response to ER stress (PubMed:26086088). Activates the transcription of asparagine synthetase (ASNS) in response to amino acid deprivation or ER stress (PubMed:11960987). However, when associated with DDIT3/CHOP, the transcriptional activation of the ASNS gene is inhibited in response to amino acid deprivation (PubMed:18940792). Together with DDIT3/CHOP, mediates programmed cell death by promoting the expression of genes involved in cellular amino acid metabolic processes, mRNA translation and the terminal unfolded protein response (terminal UPR), a cellular response that elicits programmed cell death when ER stress is prolonged and unresolved (By similarity). Activates the expression of COX7A2L/SCAF1 downstream of the EIF2AK3/PERK-mediated unfolded protein response, thereby promoting formation of respiratory chain supercomplexes and increasing mitochondrial oxidative phosphorylation (PubMed:31023583). Together with DDIT3/CHOP, activates the transcription of the IRS-regulator TRIB3 and promotes ER stress- induced neuronal cell death by regulating the expression of BBC3/PUMA in response to ER stress (PubMed:15775988). May cooperate with the UPR transcriptional regulator QRICH1 to regulate ER protein homeostasis which is critical for cell viability in response to ER stress (PubMed:33384352). In the absence of stress, ATF4 translation is at low levels and it is required for normal metabolic processes such as embryonic lens formation, fetal liver hematopoiesis, bone development and synaptic plasticity (By similarity). Acts as a regulator of osteoblast differentiation in response to phosphorylation by RPS6KA3/RSK2: phosphorylation in osteoblasts enhances transactivation activity and promotes expression of osteoblast-specific genes and post- transcriptionally regulates the synthesis of Type I collagen, the main constituent of the bone matrix (PubMed:15109498). Cooperates with FOXO1 in osteoblasts to regulate glucose homeostasis through suppression of beta-cell production and decrease in insulin production (By similarity). Activates transcription of SIRT4 (By similarity). Regulates the circadian expression of the core clock component PER2 and the serotonin transporter SLC6A4 (By similarity). Binds in a circadian time-dependent manner to the cAMP response elements (CRE) in the SLC6A4 and PER2 promoters and periodically activates the transcription of these genes (By similarity). Mainly acts as a transcriptional activator in cellular stress adaptation, but it can also act as a transcriptional repressor: acts as a regulator of synaptic plasticity by repressing transcription, thereby inhibiting induction and maintenance of long- term memory (By similarity). Regulates synaptic functions via interaction with DISC1 in neurons, which inhibits ATF4 transcription factor activity by disrupting ATF4 dimerization and DNA-binding (PubMed:31444471). |
| Cellular Location | Nucleus. Nucleus speckle. Cytoplasm {ECO:0000250|UniProtKB:Q9ES19}. Cell membrane {ECO:0000250|UniProtKB:Q9ES19}. Cytoplasm, cytoskeleton, microtubule organizing center, centrosome Note=Colocalizes with GABBR1 in hippocampal neuron dendritic membranes (By similarity). Colocalizes with NEK6 at the centrosome (PubMed:20873783). Recruited to nuclear speckles following interaction with EP300/p300 (PubMed:16219772). {ECO:0000250|UniProtKB:Q9ES19, ECO:0000269|PubMed:16219772, ECO:0000269|PubMed:20873783} |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Transcriptional activator. Binds the cAMP response element (CRE) (consensus: 5'-GTGACGT[AC][AG]-3'), a sequence present in many viral and cellular promoters. Cooperates with FOXO1 in osteoblasts to regulate glucose homeostasis through suppression of beta-cell production and decrease in insulin production (By similarity). It binds to a Tax-responsive enhancer element in the long terminal repeat of HTLV-I. Regulates the induction of DDIT3/CHOP and asparagine synthetase (ASNS) in response to ER stress. In concert with DDIT3/CHOP, activates the transcription of TRIB3 and promotes ER stress-induced neuronal apoptosis by regulating the transcriptional induction of BBC3/PUMA. Activates transcription of SIRT4. Regulates the circadian expression of the core clock component PER2 and the serotonin transporter SLC6A4. Binds in a circadian time-dependent manner to the cAMP response elements (CRE) in the SLC6A4 and PER2 promoters and periodically activates the transcription of these genes.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.