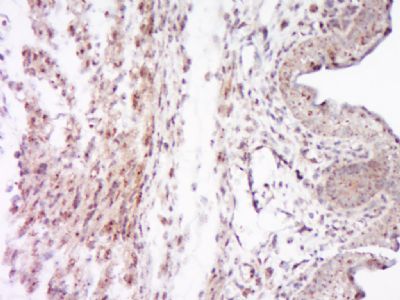
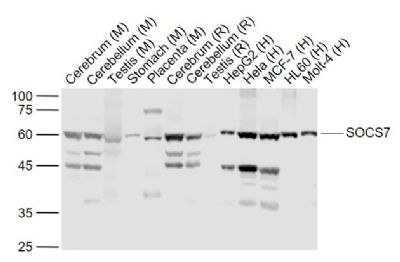
SOCS7 Polyclonal Antibody
Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IHC-F, IF, ICC, E |
|---|---|
| Primary Accession | O14512 |
| Reactivity | Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 63 KDa |
| Physical State | Liquid |
| Immunogen | KLH conjugated synthetic peptide derived from human SOCS7 |
| Epitope Specificity | 171-270/581 |
| Isotype | IgG |
| Purity | affinity purified by Protein A |
| Buffer | Preservative: 0.02% Proclin300, Constituents: 1% BSA, 0.01M PBS, pH7.4. |
| SUBCELLULAR LOCATION | Cytoplasm. Cell membrane; Peripheral membrane protein; Cytoplasmic side. Nucleus. Note=Mostly cytoplasmic, but shuttles between the cytoplasm and the nucleus. Rapidly relocalizes to the nucleus after UV irradiation. Cytoplasmic location depends upon SEPT7 presence. |
| SIMILARITY | Contains 1 SH2 domain. Contains 1 SOCS box domain. |
| SUBUNIT | Interacts with phosphorylated IRS4 and PIK3R1 (By similarity). Interacts, via the third proline-rich region, with the second SH3 domain of the adapter protein NCK1. Also interacts with GRB2, INSR, IRS1, PLCG1, SORBS3/vinexin, and phosphorylated STAT3 and STAT5. Interacts with SEPT6. |
| Important Note | This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |
| Background Descriptions | The eight members of the recently identified Suppressor of Cytokines Signaling (SOCS) family are SOCS1, SOCS2, SOCS3, SOCS4, SOCS5, SOCS6, SOCS7, and CIS. Structurally the SOCS proteins are composed of an N- terminal region of variable length and amino acid composition, a central SH2 domain, and a C-terminal motif called the SOCS box. The SOCS proteins appear to form part of a classical negative feedback loop that regulates cytokine signal transduction. Transcription of each of the SOCS genes occurs rapidly in vitro and in vivo in response to cytokines, and once produced, the various members of the SOCS family appear to inhibit signaling in different ways. SOCS1 and SOCS6 interact with the insulin receptor (IR) when expressed in human hepatoma cells (HepG2) or in rat hepatoma cells overexpressing human IR. SOCS1 and SOCS6 inhibit insulin-dependent activation of ERK1/2 and protein kinase B in vivo and IR- directed phosphorylation of IRS1 in vitro. These results suggest that SOCS proteins may be inhibitors of IR signalling and could mediate cytokine-induced insulin resistance and contribute to the pathogenesis of type II diabetes. SOCS6 and SOCS7 are expressed ubiquitously in murine tissues and SOCS6 knockout mice are growth retarded. |
| Gene ID | 30837 |
|---|---|
| Other Names | Suppressor of cytokine signaling 7, SOCS-7, Nck, Ash and phospholipase C gamma-binding protein, Nck-associated protein 4, NAP-4, SOCS7, NAP4, SOCS6 |
| Target/Specificity | Expressed in brain and leukocytes. Also in fetal lung fibroblasts and fetal brain. |
| Dilution | WB=1:500-2000,IHC-P=1:100-500,IHC-F=1:100-500,ICC=1:100-500,IF=1:100-500,ELISA=1:5000-10000 |
| Storage | Store at -20 ℃ for one year. Avoid repeated freeze/thaw cycles. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 ℃. |
| Name | SOCS7 {ECO:0000303|PubMed:16127460, ECO:0000312|HGNC:HGNC:29846} |
|---|---|
| Function | Substrate-recognition component of a cullin-5-RING E3 ubiquitin-protein ligase complex (ECS complex, also named CRL5 complex), which mediates the ubiquitination and subsequent proteasomal degradation of target proteins, such as DAB1 and IRS1 (PubMed:16127460). Specifically recognizes and binds phosphorylated proteins via its SH2 domain, promoting their ubiquitination (By similarity). The ECS(SOCS7) complex acts as a key regulator of reelin signaling by mediating ubiquitination and degradation of phosphorylated DAB1 in the cortical plate of the developing cerebral cortex, thereby regulating neuron positioning during cortex development (By similarity). Functions in insulin signaling and glucose homeostasis through IRS1 ubiquitination and subsequent proteasomal degradation (PubMed:16127460). Also inhibits prolactin, growth hormone and leptin signaling by preventing STAT3 and STAT5 activation, sequestering them in the cytoplasm and reducing their binding to DNA (PubMed:15677474). |
| Cellular Location | Cytoplasm. Nucleus Cell membrane; Peripheral membrane protein; Cytoplasmic side. Note=Mostly cytoplasmic, but shuttles between the cytoplasm and the nucleus (PubMed:17803907). Rapidly relocalizes to the nucleus after UV irradiation (PubMed:17803907) Cytoplasmic location depends upon SEPT7 presence (PubMed:17803907) |
| Tissue Location | Expressed in brain and leukocytes (PubMed:9344857). Also in fetal lung fibroblasts and fetal brain (PubMed:9344857) |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.