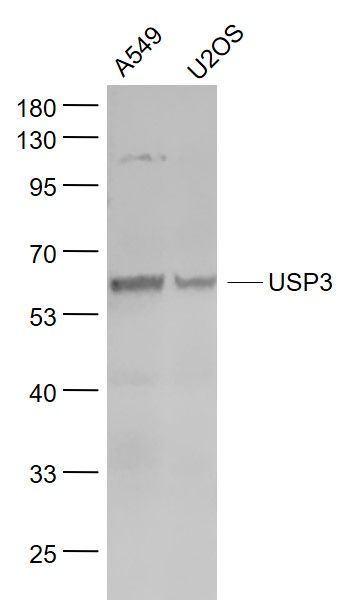
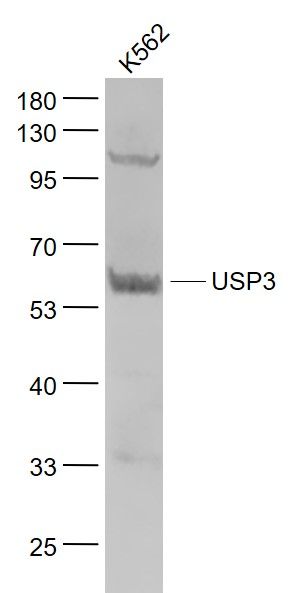
USP3 Polyclonal Antibody
Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IHC-F, IF, E |
|---|---|
| Primary Accession | Q9Y6I4 |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 59 KDa |
| Physical State | Liquid |
| Immunogen | KLH conjugated synthetic peptide derived from human USP3 |
| Epitope Specificity | 31-130/520 |
| Isotype | IgG |
| Purity | affinity purified by Protein A |
| Buffer | 0.01M TBS (pH7.4) with 1% BSA, 0.02% Proclin300 and 50% Glycerol. |
| SUBCELLULAR LOCATION | Nucleus. Localizes preferentially with monoubiquitinated H2A to chromatin. |
| SIMILARITY | Belongs to the peptidase C19 family. USP3 subfamily.Contains 1 UBP-type zinc finger. |
| SUBUNIT | Interacts (via UBP-type domain) with H2A; the interaction is less efficient than with monoubiquitinated H2A. |
| Important Note | This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |
| Background Descriptions | The ubiquitin (Ub) pathway involves three sequential enzymatic steps that facilitate the conjugation of Ub and Ub-like molecules to specific protein substrates. Through the use of a wide range of enzymes that can add or remove ubiquitin, the Ub pathway controls many intracellular processes such as signal transduction, transcriptional activation and cell cycle progression. USP3 (ubiquitin specific peptidase 3), also known as UBP or SIH003, is a 520 amino acid protein that contains one UBP-type zinc finger and belongs to the peptidase C19 family. Expressed ubiquitously with highest levels present in pancreas, USP3 catalyzes the conversion of a ubiquitin C-terminal thioester to a free ubiquitin and a thiol. The gene encoding USP3 maps to human chromosome 15, which houses over 700 genes and comprises nearly 3% of the human genome. |
| Gene ID | 9960 |
|---|---|
| Other Names | Ubiquitin carboxyl-terminal hydrolase 3, 3.4.19.12, Deubiquitinating enzyme 3, Ubiquitin thioesterase 3, Ubiquitin-specific-processing protease 3, USP3 |
| Target/Specificity | Expressed in all tissues examined, with strongest expression in pancreas. |
| Dilution | WB=1:500-2000,IHC-P=1:100-500,IHC-F=1:100-500,IF=1:50-200,ELISA=1:5000-10000 |
| Storage | Store at -20 ℃ for one year. Avoid repeated freeze/thaw cycles. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 ℃. |
| Name | USP3 |
|---|---|
| Function | Deubiquitinase that plays a role in several cellular processes including transcriptional regulation, cell cycle progression or innate immunity. In response to DNA damage, deubiquitinates monoubiquitinated target proteins such as histone H2A and H2AX and thereby counteracts RNF168- and RNF8-mediated ubiquitination. In turn, participates in the recruitment of DNA damage repair factors to DNA break sites (PubMed:24196443). Required for proper progression through S phase and subsequent mitotic entry (PubMed:17980597). Acts as a positive regulator of TP53 by deubiquitinating and stabilizing it to promote normal cell proliferation and transformation (PubMed:28807825). Participates in establishing tolerance innate immune memory through non-transcriptional feedback. Mechanistically, negatively regulates TLR-induced NF-kappa-B signaling by targeting and removing the 'Lys- 63'-linked polyubiquitin chains on MYD88 (PubMed:37971847). Negatively regulates the activation of type I interferon signaling by mediating 'Lys-63'-linked polyubiquitin chains on RIGI and IFIH1 (PubMed:24366338). Also deubiquinates ASC/PYCARD, the central adapter mediating the assembly and activation of most inflammasomes, and thereby promotes inflammasome activation (PubMed:36050480). |
| Cellular Location | Nucleus. Cytoplasm. Note=Localizes preferentially with monoubiquitinated H2A to chromatin (PubMed:17980597). Upon NF-kappa-B signaling activation, exits the nucleus (PubMed:37971847) |
| Tissue Location | Expressed in all tissues examined, with strongest expression in pancreas |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.