Histone deacetylase 6 Polyclonal Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

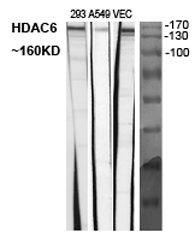
Application
| WB, IHC-P, IF |
|---|---|
| Primary Accession | Q9UBN7 |
| Reactivity | Human, Mouse |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 131419 Da |
| Gene ID | 10013 |
|---|---|
| Other Names | HDAC6; KIAA0901; JM21; Histone deacetylase 6; HD6 |
| Dilution | WB~~Western Blot: 1/500 - 1/2000. Immunohistochemistry: 1/100 - 1/300. Immunofluorescence: 1/200 - 1/1000. ELISA: 1/20000. Not yet tested in other applications. IHC-P~~N/A IF~~1:50~200 |
| Format | Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.09% (W/V) sodium azide. |
| Storage Conditions | -20℃ |
| Name | HDAC6 {ECO:0000303|PubMed:10220385, ECO:0000312|HGNC:HGNC:14064} |
|---|---|
| Function | Deacetylates a wide range of non-histone substrates (PubMed:12024216, PubMed:18606987, PubMed:20308065, PubMed:24882211, PubMed:26246421, PubMed:30538141, PubMed:31857589, PubMed:30770470, PubMed:38534334, PubMed:39567688). Plays a central role in microtubule- dependent cell motility by mediating deacetylation of tubulin (PubMed:12024216, PubMed:20308065, PubMed:26246421). Required for cilia disassembly via deacetylation of alpha-tubulin (PubMed:17604723, PubMed:26246421). Alpha-tubulin deacetylation results in destabilization of dynamic microtubules (By similarity). Promotes deacetylation of CTTN, leading to actin polymerization, promotion of autophagosome-lysosome fusion and completion of autophagy (PubMed:30538141). Deacetylates SQSTM1 (PubMed:31857589). Deacetylates peroxiredoxins PRDX1 and PRDX2, decreasing their reducing activity (PubMed:18606987). Deacetylates antiviral protein RIGI in the presence of viral mRNAs which is required for viral RNA detection by RIGI (By similarity). Sequentially deacetylates and polyubiquitinates DNA mismatch repair protein MSH2 which leads to MSH2 degradation, reducing cellular sensitivity to DNA-damaging agents and decreasing cellular DNA mismatch repair activities (PubMed:24882211). Deacetylates DNA mismatch repair protein MLH1 which prevents recruitment of the MutL alpha complex (formed by the MLH1-PMS2 heterodimer) to the MutS alpha complex (formed by the MSH2-MSH6 heterodimer), leading to tolerance of DNA damage (PubMed:30770470). Deacetylates RHOT1/MIRO1 which blocks mitochondrial transport and mediates axon growth inhibition (By similarity). Deacetylates transcription factor SP1 which leads to increased expression of ENG, positively regulating angiogenesis (PubMed:38534334). Deacetylates KHDRBS1/SAM68 which regulates alternative splicing by inhibiting the inclusion of CD44 alternate exons (PubMed:26080397). Acts as a valine sensor by binding to valine through the primate-specific SE14 repeat region (PubMed:39567688). In valine deprivation conditions, translocates from the cytoplasm to the nucleus where it deacetylates TET2 which promotes TET2-dependent DNA demethylation, leading to DNA damage (PubMed:39567688). Promotes odontoblast differentiation following IPO7-mediated nuclear import and subsequent repression of RUNX2 expression (By similarity). In addition to its protein deacetylase activity, plays a key role in the degradation of misfolded proteins: when misfolded proteins are too abundant to be degraded by the chaperone refolding system and the ubiquitin-proteasome, mediates the transport of misfolded proteins to a cytoplasmic juxtanuclear structure called aggresome (PubMed:17846173). Probably acts as an adapter that recognizes polyubiquitinated misfolded proteins and targets them to the aggresome, facilitating their clearance by autophagy (PubMed:17846173). Involved in the MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer (PubMed:24413532). |
| Cellular Location | Cytoplasm. Cytoplasm, cytoskeleton. Nucleus. Perikaryon {ECO:0000250|UniProtKB:Q9Z2V5}. Cell projection, dendrite {ECO:0000250|UniProtKB:Q9Z2V5}. Cell projection, axon {ECO:0000250|UniProtKB:Q9Z2V5}. Cell projection, cilium. Cytoplasm, cytoskeleton, microtubule organizing center, centrosome. Cytoplasm, cytoskeleton, cilium basal body Note=Mainly cytoplasmic where it is associated with microtubules (PubMed:12024216). Can shuttle between the cytoplasm and the nucleus (PubMed:39567688). Retained in the cytoplasm by binding to valine via the primate-specific SE14 repeat region while valine deprivation induces nuclear localization (PubMed:39567688). Found exclusively in the cytoplasm in proliferative cells with a fraction found in the nucleus during differentiation (By similarity). May translocate to the nucleus following DNA damage (PubMed:30770470) {ECO:0000250|UniProtKB:Q9Z2V5, ECO:0000269|PubMed:12024216, ECO:0000269|PubMed:30770470, ECO:0000269|PubMed:39567688} |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes (By similarity). Plays a central role in microtubule-dependent cell motility via deacetylation of tubulin. Involved in the MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
.jpg)

