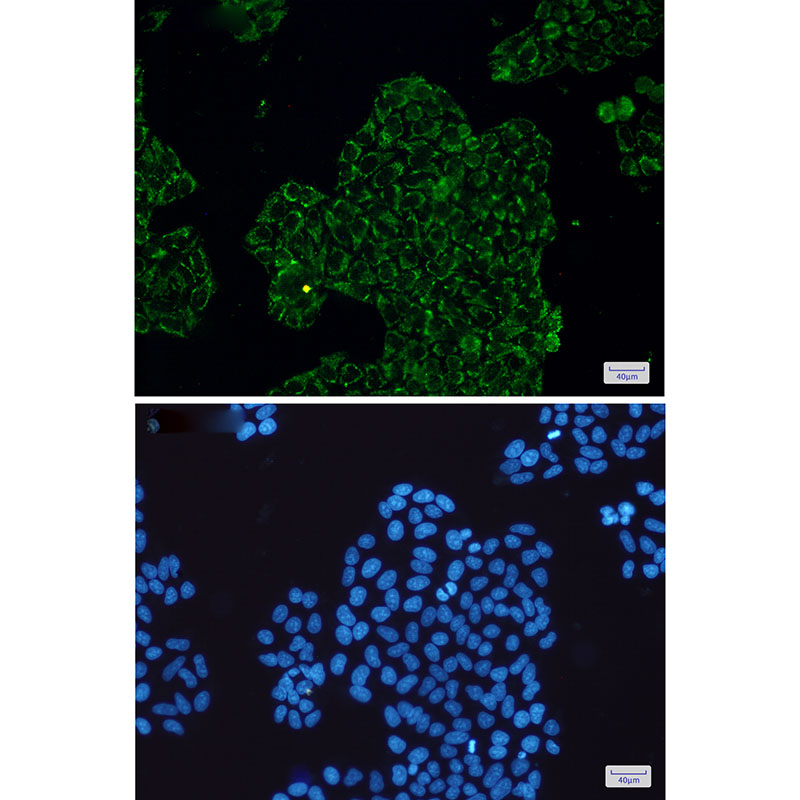
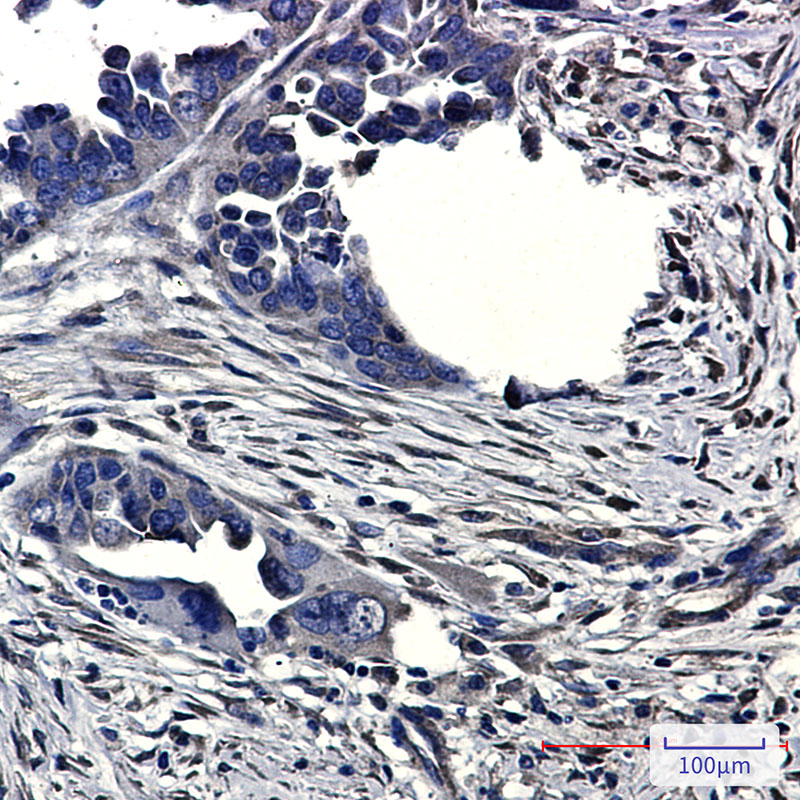
CLIC4 Rabbit mAb
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IHC-F, IP, ICC |
|---|---|
| Primary Accession | Q9Y696 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Monoclonal Antibody |
| Calculated MW | 28772 Da |
| Gene ID | 25932 |
|---|---|
| Other Names | CLIC4 |
| Dilution | WB~~1/500-1/1000 IHC-P~~N/A IHC-F~~N/A IP~~N/A ICC~~N/A |
| Format | 50mM Tris-Glycine(pH 7.4), 0.15M NaCl, 40%Glycerol, 0.01% sodium azide and 0.05% BSA. |
| Storage | Store at 4°C short term. Aliquot and store at -20°C long term. Avoid freeze/thaw cycles. |
| Name | CLIC4 {ECO:0000303|PubMed:12163372, ECO:0000312|HGNC:HGNC:13518} |
|---|---|
| Function | In the soluble state, catalyzes glutaredoxin-like thiol disulfide exchange reactions with reduced glutathione as electron donor (PubMed:25581026, PubMed:37759794). Can insert into membranes and form voltage-dependent multi-ion conductive channels. Membrane insertion seems to be redox-regulated and may occur only under oxidizing conditions (By similarity) (PubMed:16176272). Has alternate cellular functions like a potential role in angiogenesis or in maintaining apical-basolateral membrane polarity during mitosis and cytokinesis. Could also promote endothelial cell proliferation and regulate endothelial morphogenesis (tubulogenesis). Promotes cell-surface expression of HRH3. |
| Cellular Location | Cytoplasm, cytoskeleton, microtubule organizing center, centrosome. Cytoplasmic vesicle membrane; Single-pass membrane protein. Nucleus. Cell membrane; Single-pass membrane protein. Mitochondrion {ECO:0000250|UniProtKB:Q9Z0W7}. Cell junction. Endoplasmic reticulum membrane {ECO:0000250|UniProtKB:Q9Z0W7}; Single-pass membrane protein {ECO:0000250|UniProtKB:Q9Z0W7}. Note=Colocalized with AKAP9 at the centrosome and midbody. Exists both as soluble cytoplasmic protein and as membrane protein with probably a single transmembrane domain Present in an intracellular vesicular compartment that likely represent trans-Golgi network vesicles. Might not be present in the nucleus of cardiac cells. {ECO:0000250|UniProtKB:Q9Z0W7, ECO:0000269|PubMed:14569596} |
| Tissue Location | Detected in epithelial cells from colon, esophagus and kidney (at protein level). Expression is prominent in heart, kidney, placenta and skeletal muscle. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.