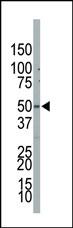
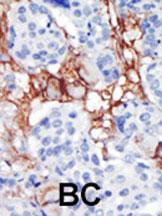
PIP5K2B Antibody (C-term)
Purified Rabbit Polyclonal Antibody (Pab)
- SPECIFICATION
- CITATIONS: 2
- PROTOCOLS
- BACKGROUND

Application
| IHC-P, WB, E |
|---|---|
| Primary Accession | P78356 |
| Other Accession | NP_003550 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | Rabbit IgG |
| Calculated MW | 47378 Da |
| Antigen Region | 286-317 aa |
| Gene ID | 8396 |
|---|---|
| Other Names | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, 1-phosphatidylinositol 5-phosphate 4-kinase 2-beta, Diphosphoinositide kinase 2-beta, Phosphatidylinositol 5-phosphate 4-kinase type II beta, PI(5)P 4-kinase type II beta, PIP4KII-beta, PtdIns(5)P-4-kinase isoform 2-beta, PIP4K2B, PIP5K2B |
| Target/Specificity | This PIP5K2B antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 286-317 amino acids from the C-terminal region of human PIP5K2B. |
| Dilution | IHC-P~~1:50~100 WB~~1:1000 E~~Use at an assay dependent concentration. |
| Format | Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is prepared by Saturated Ammonium Sulfate (SAS) precipitation followed by dialysis against PBS. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | PIP5K2B Antibody (C-term) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | PIP4K2B (HGNC:8998) |
|---|---|
| Synonyms | PIP5K2B |
| Function | Participates in the biosynthesis of phosphatidylinositol 4,5- bisphosphate (PubMed:26774281, PubMed:9038203). Preferentially utilizes GTP, rather than ATP, for PI(5)P phosphorylation and its activity reflects changes in direct proportion to the physiological GTP concentration (PubMed:26774281). Its GTP-sensing activity is critical for metabolic adaptation (PubMed:26774281). PIP4Ks negatively regulate insulin signaling through a catalytic-independent mechanism. They interact with PIP5Ks and suppress PIP5K-mediated PtdIns(4,5)P2 synthesis and insulin-dependent conversion to PtdIns(3,4,5)P3 (PubMed:31091439). |
| Cellular Location | Endoplasmic reticulum membrane; Peripheral membrane protein. Cell membrane; Peripheral membrane protein. Nucleus. Cytoplasm Note=Associated with the plasma membrane and the endoplasmic reticulum |
| Tissue Location | Highly expressed in brain, heart, pancreas, skeletal muscle and kidney. Detected at lower levels in placenta, lung and liver. |

Provided below are standard protocols that you may find useful for product applications.
Background
PIP5K2B catalyzes the phosphorylation of phosphatidylinositol-4-phosphate on the fifth hydroxyl of the myo-inositol ring to form phosphatidylinositol-4,5-bisphosphate. It is a member of the phosphatidylinositol-4-phosphate 5-kinase family. The encoded protein sequence does not show similarity to other kinases, but the protein does exhibit kinase activity. Additionally, the encoded protein interacts with p55 TNF receptor.
References
Rao, V.D., et al., Cell 94(6):829-839 (1998).
Castellino, A.M., et al., J. Biol. Chem. 272(9):5861-5870 (1997).
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.














 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.