CRISPR-Cas9 SP Antibody
Rabbit mAb
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
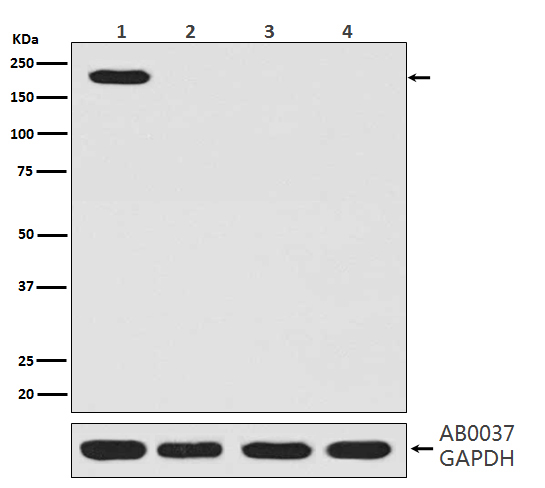
| WB, IHC, FC, ICC |
|---|---|
| Primary Accession | Q99ZW2 |
| Clonality | Monoclonal |
| Other Names | Cas9; CRISPR-associated endonuclease Cas9/Csn1; CRISPR-Cas9/Csn1; csn1; SpyCas9; |
| Isotype | Rabbit IgG |
| Host | Rabbit |
| Calculated MW | 158441 Da |
| Dilution | WB 1:1000~1:5000 IHC 1:50~1:200 ICC/IF 1:50~1:200 FC 1:50 |
|---|---|
| Purification | Affinity-chromatography |
| Immunogen | Recombinant fragment derived from Streptococcus pyogenes |
| Description | The CRISPR associated protein 9 (Cas9) is an RNA-guided DNA nuclease and part of the Streptococcus pyogenes CRISPR antiviral immunity system that provides adaptive immunity against extra chromosomal genetic material. CRISPR/Cas9 genome editing tools have been used in many organisms, including mouse and human cells. |
| Storage Condition and Buffer | Rabbit IgG in phosphate buffered saline , pH 7.4, 150mM NaCl, 0.02% sodium azide and 50% glycerol. Store at +4°C short term. Store at -20°C long term. Avoid freeze / thaw cycle. |
| Name | cas9 {ECO:0000255|HAMAP-Rule:MF_01480, ECO:0000303|PubMed:22745249} |
|---|---|
| Function | CRISPR (clustered regularly interspaced short palindromic repeat) is an adaptive immune system that provides protection against mobile genetic elements (viruses, transposable elements and conjugative plasmids) (PubMed:21455174). CRISPR clusters contain spacers, sequences complementary to antecedent mobile elements, and target invading nucleic acids. CRISPR clusters are transcribed and processed into CRISPR RNA (crRNA). In type II CRISPR systems correct processing of pre-crRNA requires a trans-encoded small RNA (tracrRNA), endogenous ribonuclease 3 (rnc) and this protein. The tracrRNA serves as a guide for ribonuclease 3-aided processing of pre-crRNA; Cas9 only stabilizes the pre-crRNA:tracrRNA interaction and has no catalytic function in RNA processing (PubMed:24270795). Subsequently Cas9/crRNA/tracrRNA endonucleolytically cleaves linear or circular dsDNA target complementary to the spacer; Cas9 is inactive in the absence of the 2 guide RNAs (gRNA). The target strand not complementary to crRNA is first cut endonucleolytically, then trimmed 3'-5' exonucleolytically. DNA-binding requires protein and both gRNAs, as does nuclease activity. Cas9 recognizes the protospacer adjacent motif (PAM) in the CRISPR repeat sequences to help distinguish self versus nonself, as targets within the bacterial CRISPR locus do not have PAMs. DNA strand separation and heteroduplex formation starts at PAM sites; PAM recognition is required for catalytic activity (PubMed:24476820). Confers immunity against a plasmid with homology to the appropriate CRISPR spacer sequences (CRISPR interference) (PubMed:21455174). |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.