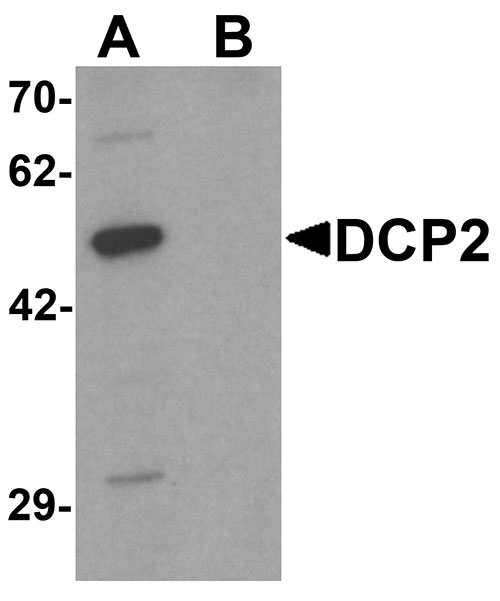
DCP2 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IF, E |
|---|---|
| Primary Accession | Q8IU60 |
| Other Accession | NP_689837, 31542498 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Calculated MW | 46 kDa |
| Application Notes | DCP2 antibody can be used for detection of DCP2 by Western blot at 1 µg/mL. For immunofluorescence start at 20 µg/mL. |
| Gene ID | 167227 |
|---|---|
| Target/Specificity | DCP2; Multiple isoforms of DCP2 are known to exist. |
| Reconstitution & Storage | DCP2 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. As with all antibodies care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. |
| Precautions | DCP2 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | DCP2 |
|---|---|
| Synonyms | NUDT20 |
| Function | Decapping metalloenzyme that catalyzes the cleavage of the cap structure on mRNAs (PubMed:12218187, PubMed:12417715, PubMed:12923261, PubMed:21070968, PubMed:28002401, PubMed:31875550). Removes the 7-methyl guanine cap structure from mRNA molecules, yielding a 5'-phosphorylated mRNA fragment and 7m-GDP (PubMed:12486012, PubMed:12923261, PubMed:21070968, PubMed:28002401, PubMed:31875550). Necessary for the degradation of mRNAs, both in normal mRNA turnover and in nonsense-mediated mRNA decay (PubMed:14527413). Plays a role in replication-dependent histone mRNA degradation (PubMed:18172165). Has higher activity towards mRNAs that lack a poly(A) tail (PubMed:21070968). Has no activity towards a cap structure lacking an RNA moiety (PubMed:21070968). The presence of a N(6)-methyladenosine methylation at the second transcribed position of mRNAs (N(6),2'-O- dimethyladenosine cap; m6A(m)) provides resistance to DCP2-mediated decapping (PubMed:28002401). Blocks autophagy in nutrient-rich conditions by repressing the expression of ATG-related genes through degradation of their transcripts (PubMed:26098573). |
| Cellular Location | Cytoplasm, P-body. Nucleus Note=Predominantly cytoplasmic, in processing bodies (PB) (PubMed:15273322). A minor amount is nuclear (PubMed:15273322) |
| Tissue Location | Expressed in brain and testis. Not detected in heart (at protein level). |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
DCP2 Antibody: The removal, or decapping, of eukaryotic mRNA is an important step in the degradation of mRNA. Decapping protein 2 (DCP2) is the major mRNA decapping enzyme in cells. It is a member of the Nudix hydrolases superfamily of proteins that predominantly catalyze the hydrolysis of small nucleoside diphosphate substrates linked to another moiety. DCP2 is widely expressed in multiple tissues at varying levels, with highest expression seen in testis and brain.
References
Song MG, Li Y, and Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol. Cell 2010; 40:423-32.
Dunckley T and Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411-22.
Bessman MJ, Frick DN, and O’Handley SF. The MutT proteins of “Nudix" hydrolases, a family of versatile, widely distributed, “housecleaning" enzymes. J. Biol. Chem. 1996; 271:25059-62
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.