UNG2 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IF, E |
|---|---|
| Primary Accession | P13051 |
| Other Accession | NP_550433, 19718751 |
| Reactivity | Human, Mouse |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
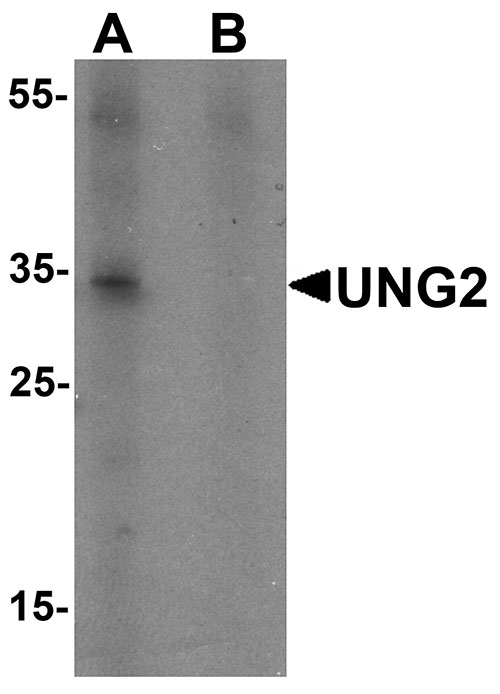
| Calculated MW | Predicted: 34 kDa Observed: 33 kDa |
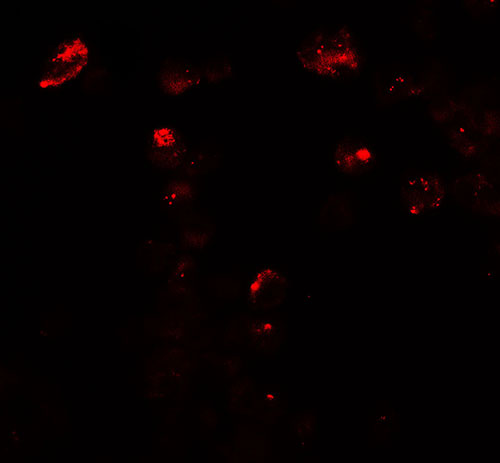
| Application Notes | UNG2 antibody can be used for detection of UNG2 by Western blot at 1 - 2 µg/mL. For immunofluorescence start at 20 µg/mL. |
| Gene ID | 7374 |
|---|---|
| Target/Specificity | UNG; At least two isoforms of UNG2 are known to exist; this antibody will only detect the longer isoform. UNG2 antibody is predicted to not cross-react with UNG1. |
| Reconstitution & Storage | UNG2 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. |
| Precautions | UNG2 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | UNG {ECO:0000255|HAMAP-Rule:MF_03166} |
|---|---|
| Function | Uracil-DNA glycosylase that hydrolyzes the N-glycosidic bond between uracil and deoxyribose in single- and double-stranded DNA (ssDNA and dsDNA) to release a free uracil residue and form an abasic (apurinic/apyrimidinic; AP) site. Excises uracil residues arising as a result of misincorporation of dUMP residues by DNA polymerase during replication or due to spontaneous or enzymatic deamination of cytosine (PubMed:12958596, PubMed:15967827, PubMed:17101234, PubMed:22521144, PubMed:7671300, PubMed:8900285, PubMed:9016624, PubMed:9776759). Mediates error-free base excision repair (BER) of uracil at replication forks. According to the model, it is recruited by PCNA to S-phase replication forks to remove misincorporated uracil at U:A base mispairs in nascent DNA strands. Via trimeric RPA it is recruited to ssDNA stretches ahead of the polymerase to allow detection and excision of deaminated cytosines prior to replication. The resultant AP sites temporarily stall replication, allowing time to repair the lesion (PubMed:22521144). Mediates mutagenic uracil processing involved in antibody affinity maturation. Processes AICDA-induced U:G base mispairs at variable immunoglobulin (Ig) regions leading to the generation of transversion mutations (PubMed:12958596). Operates at switch sites of Ig constant regions where it mediates Ig isotype class switch recombination. Excises AICDA-induced uracil residues forming AP sites that are subsequently nicked by APEX1 endonuclease. The accumulation of staggered nicks in opposite strands results in double strand DNA breaks that are finally resolved via non-homologous end joining repair pathway (By similarity) (PubMed:12958596). |
| Cellular Location | [Isoform 1]: Mitochondrion |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
UNG2 Antibody: The human uracil-DNA glycosylase (UNG) gene encodes both mitochondrial (UNG1) and nuclear (UNG2) forms through differentially regulated promoters and alternative splicing. UNG2 is the major enzyme in the base excision repair pathway that removes uracil residues from DNA that arise through either misincorporation during replication or cytosine deamination. UNG2 can also be bound by the HIV-1 integrase and incorporated into the virion particle, suggesting that it is required to remove uracils from the viral genome. As the intrinsic antiviral protein APOBEC3G generates numerous uracils in the HIV genome during its replication, it may be that the UNG2 contributes to the APOBEC3G-mediated loss of infectivity by generating abasic sites in the viral genome.
References
Krokan HE, Otterlei M, Nilsen H, et al. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog. Nucleic Acid Res. Mol. Biol. 2001; 68:365-86.
Fromm JC and Verdine GL. Base excision repair. Adv. Protein Chem. 2004; 69:1-41.
Willetts KE, Rey F, Agostini I, et al. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J. Virol. 1999; 73:1682-8.
Harris RS, Bishop KN, Sheehy AM, et al. DNA deamination mediates innate immunity to retroviral infection. Cell 2003; 113:803-9.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.