AIMP2 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IF, E |
|---|---|
| Primary Accession | Q13155 |
| Other Accession | NP_006294, 11125770 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
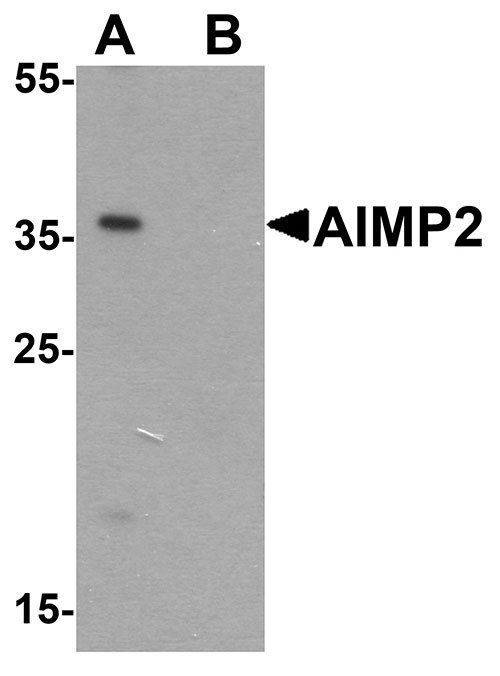
| Calculated MW | Predicted: 35 kDa Observed: 36 kDa |
| Application Notes | AIMP2 antibody can be used for detection of AIMP2 by Western blot at 1 - 2 µg/ml. |
| Gene ID | 7965 |
|---|---|
| Target/Specificity | AIMP2; AIMP2 antibody is human specific. AIMP2 antibody is predicted to not cross-react with AIMP1. |
| Reconstitution & Storage | AIMP2 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. |
| Precautions | AIMP2 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | AIMP2 |
|---|---|
| Synonyms | JTV1 |
| Function | Required for assembly and stability of the aminoacyl-tRNA synthase complex (PubMed:19131329). Mediates ubiquitination and degradation of FUBP1, a transcriptional activator of MYC, leading to MYC down-regulation which is required for aveolar type II cell differentiation. Blocks MDM2-mediated ubiquitination and degradation of p53/TP53. Functions as a proapoptotic factor. |
| Cellular Location | Cytoplasm, cytosol. Nucleus {ECO:0000250|UniProtKB:Q8R010}. Note=Following DNA damage, dissociates from the aminoacyl-tRNA synthase complex and translocates from the cytoplasm to the nucleus. {ECO:0000250|UniProtKB:Q8R010} |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
AIMP2 was initially identified as a part of an aminoacyl-tRNA synthesase complex (1). It was later discovered to be a cofactor and substrate of Parkin, a Ring-type E3 ubiquitin ligase that is important for the survival of dopamine neurons in Parkinson’s disease; accumulation of AIMP2 in these cells lead to catecholaminergic cell death (2). AIMP2 can also bind to TRAF2, a key player in the TNF-alpha signaling pathway, causing the ubiquitination of TRAF2 by cIAP1, leading to TNF-alpha-dependent apoptosis (3). Finally, AIMP2 has been suggested to function as a tumor suppressor (4).
References
Quevillon S, Robinson JC, Berthonneau E, et al. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 1999; 285:183-95.
Ko HS, von Coelln R, Sriram SR, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neruosci. 2005; 25:7968-78.
Choi JW, Kim DG, Park MC, et al. AIMP2 promotes TNFalpha-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J. Cell Sci. 2009; 122:2710-5.
Choi JW, UM JY, Kundu JK, et al. Multidirectional tumor-suppressive activity of AIMP2/p38 and the enhanced susceptibility of AIMP2 heterozygous mice to carcinogenesis. Carcinogenesis 2009; 30:1638-44.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.