SUZ12 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IF, E |
|---|---|
| Primary Accession | Q15022 |
| Other Accession | NP_056170, 197333809 |
| Reactivity | Human |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
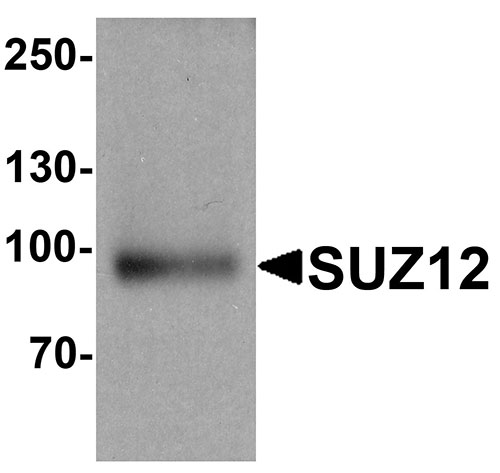
| Calculated MW | Predicted: 81 kDa Observed: 96 kDa |
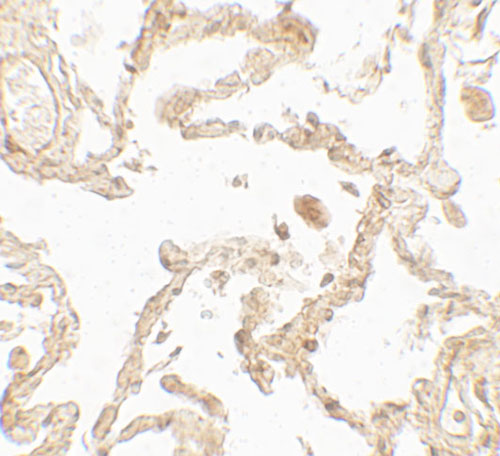
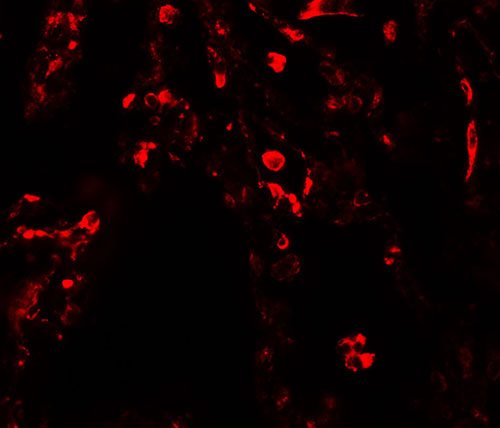
| Application Notes | SUZ12 antibody can be used for detection of SUZ12 by Western blot at 1 - 2 µg/ml. Antibody can also be used for Immunohistochemistry starting at 5 µg/mL. For immunofluorescence start at 20 µg/mL. |
| Gene ID | 23512 |
|---|---|
| Target/Specificity | SUZ12; SUZ12 antibody is human specific. |
| Reconstitution & Storage | SUZ12 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. |
| Precautions | SUZ12 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | SUZ12 |
|---|---|
| Synonyms | CHET9, JJAZ1, KIAA0160 |
| Function | Polycomb group (PcG) protein. Component of the PRC2 complex, which methylates 'Lys-9' (H3K9me) and 'Lys-27' (H3K27me) of histone H3, leading to transcriptional repression of the affected target gene (PubMed:15225548, PubMed:15231737, PubMed:15385962, PubMed:16618801, PubMed:17344414, PubMed:18285464, PubMed:28229514, PubMed:29499137, PubMed:31959557). The PRC2 complex may also serve as a recruiting platform for DNA methyltransferases, thereby linking two epigenetic repression systems (PubMed:12351676, PubMed:12435631, PubMed:15099518, PubMed:15225548, PubMed:15385962, PubMed:15684044, PubMed:16431907, PubMed:18086877, PubMed:18285464). Genes repressed by the PRC2 complex include HOXC8, HOXA9, MYT1 and CDKN2A (PubMed:15231737, PubMed:16618801, PubMed:17200670, PubMed:31959557). |
| Cellular Location | Nucleus Note=Localizes to chromatin as part of the PRC2 complex |
| Tissue Location | Overexpressed in breast and colon cancer. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
The Polycomb group (PcG) genes contribute to the maintenance of cell identity, cell cycle regulation and oncogenesis (1). SUZ12 (suppressor of zeste 12) is a 90 - 95 kDa member of the polycomb group of transcriptional regulators (2). It is part of a transcription repression complex termed PRC2 (Polycomb repressive complex 2) that methylates histones in the nucleus, resulting in homeotic (pattern-inducing) gene silencing. It contains one C2H2-type zinc finger region and a C-terminal VEFS-box (3,4). SUZ12 is overexpressed in breast and colon cancer. SUZ12 may be a cause of endometrial stromal tumors (5).
References
Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of Histone H3 Lys 27. Genes Dev. 2004; 18:1592-605.
Pasini D, Bracken AP, Hansen JB, et al. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007; 27:3769-79.
Cao R and Zhang Y. SUZ12 is required for both the Histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004; 15:57-67.
Pasini D, Bracken AP, Jensen MR, et al. SUZ12 is essential for mouse development and for EZH2 Histone methyltransferase activity. EMBO J. 2004; 23:4061-71.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.