MIB1 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IF, ICC, E |
|---|---|
| Primary Accession | Q86YT6 |
| Other Accession | NP_065825, 30348954 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
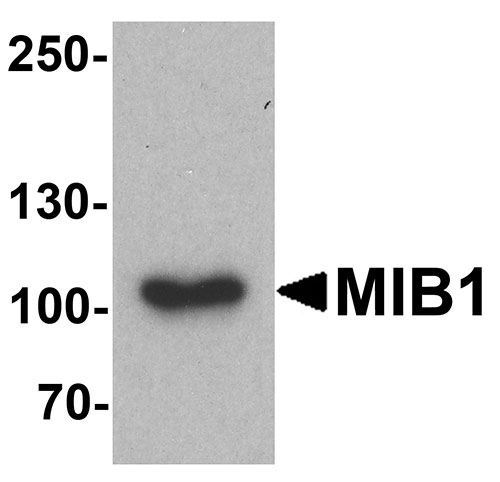
| Calculated MW | Predicted: 89, 102, 111 kDa Observed: 103 kDa |
| Application Notes | MIB1 antibody can be used for detection of MIB1 by Western blot at 1 - 2 µg/ml. Antibody can also be used for Immunocytochemistry at 5 µg/mL. For Immunoflorescence start at 20 µg/mL. |
| Gene ID | 57534 |
|---|---|
| Target/Specificity | MIB1; MIB1 antibody is human, mouse and rat reactive. At least three isoforms of MIB1 are known to exist. |
| Reconstitution & Storage | MIB1 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. |
| Precautions | MIB1 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | MIB1 |
|---|---|
| Synonyms | DIP1, KIAA1323, ZZANK2 |
| Function | E3 ubiquitin-protein ligase that mediates ubiquitination of Delta receptors, which act as ligands of Notch proteins. Positively regulates the Delta-mediated Notch signaling by ubiquitinating the intracellular domain of Delta, leading to endocytosis of Delta receptors. Probably mediates ubiquitination and subsequent proteasomal degradation of DAPK1, thereby antagonizing anti-apoptotic effects of DAPK1 to promote TNF-induced apoptosis (By similarity). Involved in ubiquitination of centriolar satellite CEP131, CEP290 and PCM1 proteins and hence inhibits primary cilium formation in proliferating cells. Mediates 'Lys-63'-linked polyubiquitination of TBK1, which probably participates in kinase activation. |
| Cellular Location | Cytoplasm. Cytoplasm, cytoskeleton, microtubule organizing center, centrosome, centriolar satellite. Cell membrane. Note=Localizes to the plasma membrane (By similarity) According to PubMed:15048887, it is mitochondrial, however such localization remains unclear. Displaced from centriolar satellites in response to cellular stress, such as ultraviolet light (UV) radiation or heat shock. |
| Tissue Location | Widely expressed at low level. Expressed at higher level in spinal cord, ovary, whole brain, and all specific brain regions examined. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
The E3 ubiquitin-protein ligase MIB1, also known as Mindbomb homolog 1, contains multiple ankyrin repeats and RING finger domains that functions as an E3 ubiquitin ligase (1). MIB1 interacts with and regulates the cellular expression of the death-associated protein kinase 1 (DAPK1) protein by promoting its ubiquitination and degradation (1). It also positively regulates Notch signaling by ubiquitinating the Notch receptors, thereby facilitating their endocytosis (2).
References
Jin Y, Blue EK, Dixon S, et al. A death-associated protein kinase (DAPK)-interacting protein, DIP-1, is an E3 ubiquitin ligase that promotes tumor necrosis factor-induced apoptosis and regulates the cellular level of DAPK. J. Biol. Chem. 2002; 277:46980-6.
Hansson EM, Lanner F, Das D, et al. Control of Notch0ligand endocytosis by ligand-receptor interaction. J. Cell Sci. 2010; 123:2931-42.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.