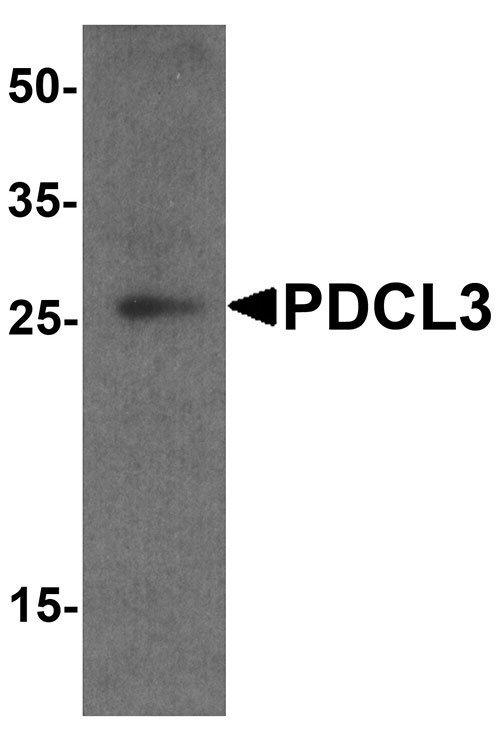
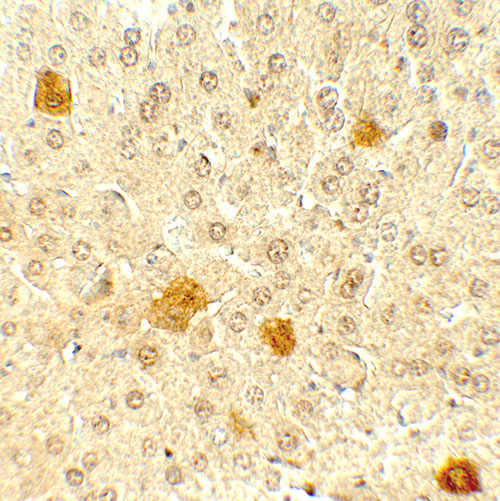
PDCL3 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IF, E |
|---|---|
| Primary Accession | Q9H2J4 |
| Other Accession | NP_076970, 13129044 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Calculated MW | Predicted: 26 kDa Observed: 25 kDa |
| Application Notes | PDCL3 antibody can be used for detection of PDCL by Western blot at 1 - 2 µg/ml. Antibody can also be used for Immunohistochemistry at 5 µg/mL. For Immunoflorescence start at 20 µg/mL. |
| Gene ID | 79031 |
|---|---|
| Target/Specificity | PDCL3; PDCL3 antibody is human, mouse and rat reactive. PDCL3 antibody is predicted to not cross-react with other members of the PDCL protein family. |
| Reconstitution & Storage | PDCL3 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. |
| Precautions | PDCL3 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | PDCL3 |
|---|---|
| Synonyms | PhLP2A, VIAF1 |
| Function | Acts as a chaperone for the angiogenic VEGF receptor KDR/VEGFR2, increasing its abundance by inhibiting its ubiquitination and degradation (PubMed:23792958, PubMed:26059764). Inhibits the folding activity of the chaperonin-containing T-complex (CCT) which leads to inhibition of cytoskeletal actin folding (PubMed:17429077). Acts as a chaperone during heat shock alongside HSP90 and HSP40/70 chaperone complexes (By similarity). Modulates the activation of caspases during apoptosis (PubMed:15371430). |
| Cellular Location | Cytoplasm. Cytoplasm, perinuclear region. Endoplasmic reticulum |
| Tissue Location | Expressed in endothelial cells (at protein level) (PubMed:26059764). Expressed in all tissues examined including spleen, thymus, prostate, testis, ovary, small intestine and colon (PubMed:15371430). |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Phosducin-like proteins (PhLPs) are a conserved family of proteins with thioredoxin-like domains that were initially identified as modulators of G protein signaling (1,2). PDCL3 is highly homologous to PDCL and shares an N-terminal helix domain and a C-terminal thioredoxin-fold (Trx-fold) domain (3). Along with the related protein PDCL2, PDCL3 interacts with the chaperonin CCT and modulates CCT-mediated actin and tubulin folding (4). Modulation of PDCL3 levels by MAPK phosphorylation and RhoA-dependent changes also promote cytoskeletal remodeling (5).
References
Miles MF, Barhite S, Sganga M, et al. Phosducin-like protein: an ethanol-responsive potential modulator of guanine nucleotide-binding protein function. Proc. Natl. Acad. Sci. USA 1993; 90:10831-5.
Ruiz-Gomez A, Humrich J, Murga C, et al. Phosphorylation of phosducin and phosducinlike protein by G protein-coupled receptor kinase 2. J. Biol. Chem. 2000; 275:29724-30.
Lou X, Bao R, Zhou CZ, et al. Structure of the thioredoxin-fold domain of human phosducin-like protein 2. Acta Crystallographica 2009; F65:67-70.
Stirling PC, Srayko M, Takhar KS, et al. Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeltal protein function and cell cycle progrssion. Mol. Biol. Cell 2007; 18:2336-45.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.