HDAC2 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IF, E |
|---|---|
| Primary Accession | Q92769 |
| Other Accession | NP_001518, 293336691 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
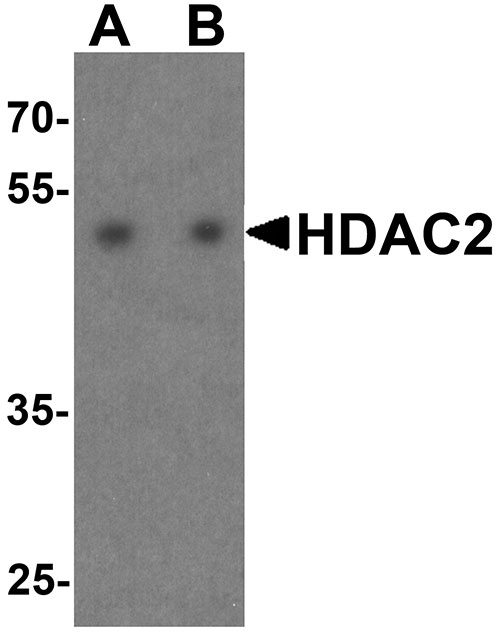
| Calculated MW | Predicted: 54 kDa Observed: 52 kDa |
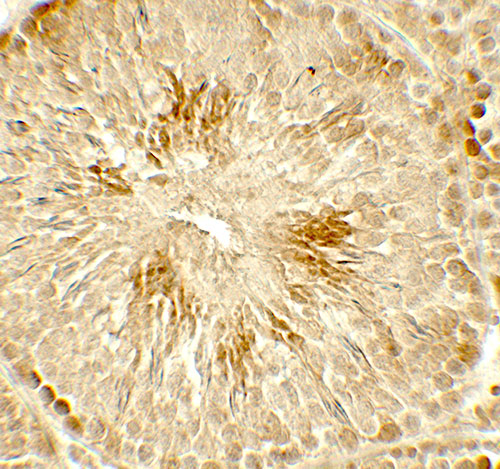
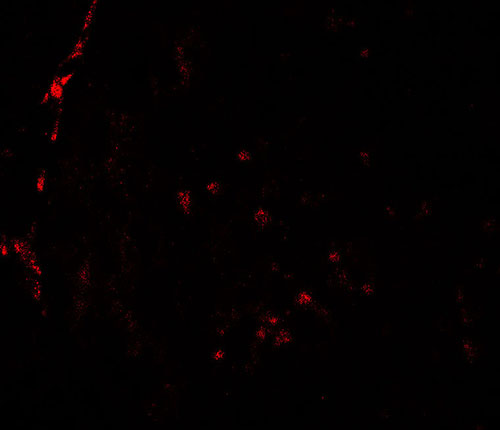
| Application Notes | HDAC2 antibody can be used for detection of HDAC2 by Western blot at 0.5 - 1 µg/ml. Antibody can also be used for Immunohistochemistry at 5 µg/mL. For Immunoflorescence start at 20 µg/mL. |
| Gene ID | 3066 |
|---|---|
| Target/Specificity | HDAC2; HDAC2 antibody is human, mouse and rat reactive. At least two isoforms of HDAC2 are known to exist; this antibody will detect both isoforms. HDAC2 antibody is predicted to not cross-react with other members of the HDAC family. |
| Reconstitution & Storage | HDAC2 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. |
| Precautions | HDAC2 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | HDAC2 {ECO:0000303|PubMed:10545197, ECO:0000312|HGNC:HGNC:4853} |
|---|---|
| Function | Histone deacetylase that catalyzes the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4) (PubMed:28497810). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events (By similarity). Histone deacetylases act via the formation of large multiprotein complexes (By similarity). Forms transcriptional repressor complexes by associating with MAD, SIN3, YY1 and N-COR (PubMed:12724404). Component of a RCOR/GFI/KDM1A/HDAC complex that suppresses, via histone deacetylase (HDAC) recruitment, a number of genes implicated in multilineage blood cell development (By similarity). Acts as a component of the histone deacetylase NuRD complex which participates in the remodeling of chromatin (PubMed:16428440, PubMed:28977666). Component of the SIN3B complex that represses transcription and counteracts the histone acetyltransferase activity of EP300 through the recognition H3K27ac marks by PHF12 and the activity of the histone deacetylase HDAC2 (PubMed:37137925). Also deacetylates non-histone targets: deacetylates TSHZ3, thereby regulating its transcriptional repressor activity (PubMed:19343227). May be involved in the transcriptional repression of circadian target genes, such as PER1, mediated by CRY1 through histone deacetylation (By similarity). Involved in MTA1-mediated transcriptional corepression of TFF1 and CDKN1A (PubMed:21965678). In addition to protein deacetylase activity, also acts as a protein-lysine deacylase by recognizing other acyl groups: catalyzes removal of (2E)-butenoyl (crotonyl), lactoyl (lactyl) and 2-hydroxyisobutanoyl (2-hydroxyisobutyryl) acyl groups from lysine residues, leading to protein decrotonylation, delactylation and de-2-hydroxyisobutyrylation, respectively (PubMed:28497810, PubMed:29192674, PubMed:35044827). |
| Cellular Location | Nucleus. Cytoplasm |
| Tissue Location | Widely expressed; lower levels in brain and lung. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
The histone deacetylase (HDAC) family contains multiple members which are divided into four classes. Class I of the HDAC family comprises four members, HDAC1, 2, 3, and 8, each of which contains a deacetylase domain and exhibits a different, individual substrate specificity and function in vivo (1). HDAC2 was first identified as as a mammalian homolog to the yeast transcriptional regulator RPD3 (2). HDAC2 forms transcriptional repressor complexes by associating with many different proteins, including YY1, a mammalian zinc-finger transcription factor (2,3). Thus, it plays an important role in transcriptional regulation, cell cycle progression and developmental events (4).
References
Taunton J, Hassig CA, and Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996; 272:408-11.
Yang WM, Inouye C, Zeng Y, et al. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 1996; 93:12845-50.
Cress WD and Seto E. Histone deacetylases, transcriptional control, and cancer. J. Cell Phys. 2000; 184:1-16.
Kramer OH. HDAC2: a critical factor in health and disease. Trends Pharmacol. Sci. 2009; 30:647-55.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.