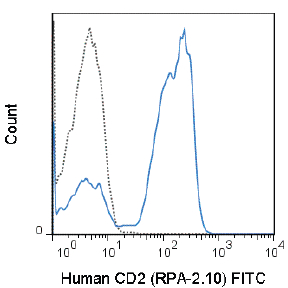
FITC Anti-Human CD2 (RPA-2.10) Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| FC |
|---|---|
| Isotype | Mouse IgG1, kappa |
| Concentration | 5 µL (0.25 µg)/test |
| Reactivity | Human |
| Formulation | 10 mM NaH2PO4, 150 mM NaCl, 0.09% NaN3, 0.1% gelatin, pH7.2 |
| Gene ID | 914 |
|---|---|
| Gene Name | CD2 |
| Alternative Name(s) | LFA-2, T11, Sheep Red Blood Cell Receptor (SRBC-R), Ly-37 |
| Format | FITC |
| Storage Conditions | 2-8°C protected from light |
Provided below are standard protocols that you may find useful for product applications.
Background
The RPA-2.10 antibody reacts with human CD2, an approximately 50 kDa glycoprotein, and a member of the Ig superfamily. CD2, also known as LFA-2, is a receptor for CD58 in the human and is expressed on the cell surface of 80-90% of human peripheral blood lymphocytes, a subset of NK cells, and all mature T cells. CD2 mediates lymphocyte adhesion and is involved in T cell activation. RPA-2.10 is reported to block mixed lymphocyte reaction. Please choose the appropriate format for each application
References
Knapp W, Dorken B, et al. eds. 1989. Leucocyte Typing IV: White Cell Differentiation Antigens. Oxford University Press. New York.
Aversa GG, Bishop GA, Suranyi MG and Hall BM. 1987. Transplant Proc. 19: 277-278.
Hahn WC, Burakoff SJ, Bierer BE. 1993. J Immunol. 150(7):2607-2619.
Xu H, Elster EA, Blair PJ, Burkly LC, Tadaki DK, Harlan DM and Kirk AD. 2003. Am J Transplant. 3: 1350-1354. (Immunohistochemistry – frozen tissue)
Kim EO, Kim TJ, Kim N, Kim ST, Kumar V and Lee KM. 2010. J Biol Chem. 285: 41755-41764. (in vitro blocking)
Schernthaner GH, Jordan JH, Ghannadan M, Agis H, Bevec D, Nunez R, Escribano L, Majdic O, Willheim M, Worda C, Printz D, Fritsch G, Lechner K and Valent P. 2001. Blood. 98(13): 3784-3792. (Flow Cytometry)
Herodin F, Thullier P, Garin D and Drouet M. 2005. Eur Cytokine Netw. 16(2): 104-116. (Flow Cytometry – Baboon, Chimpanzee, Cynomolgus, Rhesus)
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.














 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.