Anti-SIRT5/Sirtuin 5 Rabbit Monoclonal Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

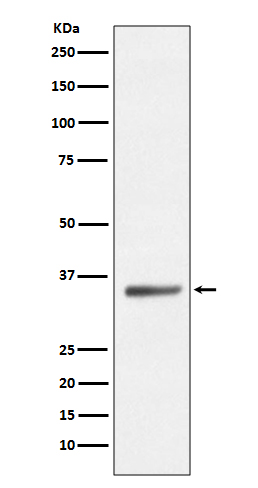
Application
| WB |
|---|---|
| Primary Accession | Q9NXA8 |
| Host | Rabbit |
| Isotype | Rabbit IgG |
| Reactivity | Human, Mouse |
| Clonality | Monoclonal |
| Format | Liquid |
| Description | Anti-SIRT5/Sirtuin 5 Rabbit Monoclonal Antibody . Tested in WB application. This antibody reacts with Human, Mouse. |
| Gene ID | 23408 |
|---|---|
| Other Names | NAD-dependent protein deacylase sirtuin-5, mitochondrial {ECO:0000255|HAMAP-Rule:MF_03160}, 2.3.1.- {ECO:0000255|HAMAP-Rule:MF_03160, ECO:0000269|PubMed:22076378, ECO:0000269|PubMed:24703693, ECO:0000269|PubMed:29180469}, Regulatory protein SIR2 homolog 5 {ECO:0000255|HAMAP-Rule:MF_03160}, SIR2-like protein 5 {ECO:0000255|HAMAP-Rule:MF_03160}, SIRT5 {ECO:0000255|HAMAP-Rule:MF_03160}, SIR2L5 |
| Calculated MW | 33881 MW KDa |
| Application Details | WB 1:1000-1:2000 |
| Subcellular Localization | Mitochondrion matrix. Mitochondrion intermembrane space. Cytoplasm, cytosol. Nucleus. Mainly mitochondrial. Also present extramitochondrially: a fraction is present in the cytosol and very small amounts are also detected in the nucleus. |
| Tissue Specificity | Widely expressed.. |
| Contents | Rabbit IgG in phosphate buffered saline, pH 7.4, 150mM NaCl, 0.02% sodium azide and 50% glycerol, 0.4-0.5mg/ml BSA. |
| Clone Names | Clone: HHE-19 |
| Immunogen | A synthesized peptide derived from human SIRT5 |
| Purification | Affinity-chromatography |
| Storage | Store at -20°C for one year. For short term storage and frequent use, store at 4°C for up to one month. Avoid repeated freeze-thaw cycles. |
| Name | SIRT5 {ECO:0000255|HAMAP-Rule:MF_03160} |
|---|---|
| Synonyms | SIR2L5 |
| Function | NAD-dependent lysine demalonylase, desuccinylase and deglutarylase that specifically removes malonyl, succinyl and glutaryl groups on target proteins (PubMed:21908771, PubMed:22076378, PubMed:24703693, PubMed:29180469). Activates CPS1 and contributes to the regulation of blood ammonia levels during prolonged fasting: acts by mediating desuccinylation and deglutarylation of CPS1, thereby increasing CPS1 activity in response to elevated NAD levels during fasting (PubMed:22076378, PubMed:24703693). Activates SOD1 by mediating its desuccinylation, leading to reduced reactive oxygen species (PubMed:24140062). Activates SHMT2 by mediating its desuccinylation (PubMed:29180469). Modulates ketogenesis through the desuccinylation and activation of HMGCS2 (By similarity). Has weak NAD-dependent protein deacetylase activity; however this activity may not be physiologically relevant in vivo. Can deacetylate cytochrome c (CYCS) and a number of other proteins in vitro such as UOX. |
| Cellular Location | Mitochondrion matrix. Mitochondrion intermembrane space. Cytoplasm, cytosol. Nucleus. Note=Mainly mitochondrial. Also present extramitochondrially, with a fraction present in the cytosol and very small amounts also detected in the nucleus [Isoform 2]: Mitochondrion {ECO:0000255|HAMAP- Rule:MF_03160, ECO:0000269|PubMed:21143562} |
| Tissue Location | Widely expressed.. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.