Hrs (phospho Tyr216) Polyclonal Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

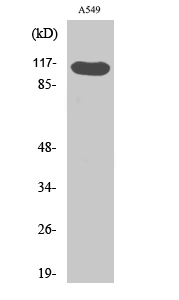
Application
| WB |
|---|---|
| Primary Accession | O14964 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 86192 Da |
| Gene ID | 9146 |
|---|---|
| Other Names | HGS; HRS; Hepatocyte growth factor-regulated tyrosine kinase substrate; Hrs; Protein pp110 |
| Dilution | WB~~Western Blot: 1/500 - 1/2000. ELISA: 1/10000. Not yet tested in other applications. |
| Format | Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.09% (W/V) sodium azide. |
| Storage Conditions | -20℃ |
| Name | HGS |
|---|---|
| Synonyms | HRS |
| Function | Involved in intracellular signal transduction mediated by cytokines and growth factors. When associated with STAM, it suppresses DNA signaling upon stimulation by IL-2 and GM-CSF. Could be a direct effector of PI3-kinase in vesicular pathway via early endosomes and may regulate trafficking to early and late endosomes by recruiting clathrin. May concentrate ubiquitinated receptors within clathrin- coated regions. Involved in down-regulation of receptor tyrosine kinase via multivesicular body (MVBs) when complexed with STAM (ESCRT-0 complex). The ESCRT-0 complex binds ubiquitin and acts as a sorting machinery that recognizes ubiquitinated receptors and transfers them to further sequential lysosomal sorting/trafficking processes. May contribute to the efficient recruitment of SMADs to the activin receptor complex. Involved in receptor recycling via its association with the CART complex, a multiprotein complex required for efficient transferrin receptor recycling but not for EGFR degradation. |
| Cellular Location | Cytoplasm {ECO:0000250|UniProtKB:Q9JJ50}. Early endosome membrane; Peripheral membrane protein; Cytoplasmic side Endosome, multivesicular body membrane {ECO:0000250|UniProtKB:Q9JJ50}; Peripheral membrane protein {ECO:0000250|UniProtKB:Q9JJ50} Note=Colocalizes with UBQLN1 in ubiquitin-rich cytoplasmic aggregates that are not endocytic compartments. |
| Tissue Location | Ubiquitous expression in adult and fetal tissues with higher expression in testis and peripheral blood leukocytes |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Involved in intracellular signal transduction mediated by cytokines and growth factors. When associated with STAM, it suppresses DNA signaling upon stimulation by IL-2 and GM-CSF. Could be a direct effector of PI3-kinase in vesicular pathway via early endosomes and may regulate trafficking to early and late endosomes by recruiting clathrin. May concentrate ubiquitinated receptors within clathrin-coated regions. Involved in down- regulation of receptor tyrosine kinase via multivesicular body (MVBs) when complexed with STAM (ESCRT-0 complex). The ESCRT-0 complex binds ubiquitin and acts as sorting machinery that recognizes ubiquitinated receptors and transfers them to further sequential lysosomal sorting/trafficking processes. May contribute to the efficient recruitment of SMADs to the activin receptor complex. Involved in receptor recycling via its association with the CART complex, a multiprotein complex required for efficient transferrin receptor recycling but not for EGFR degradation.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.