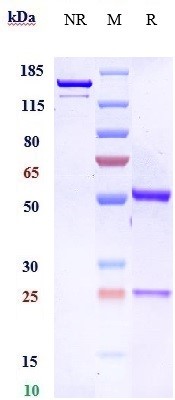
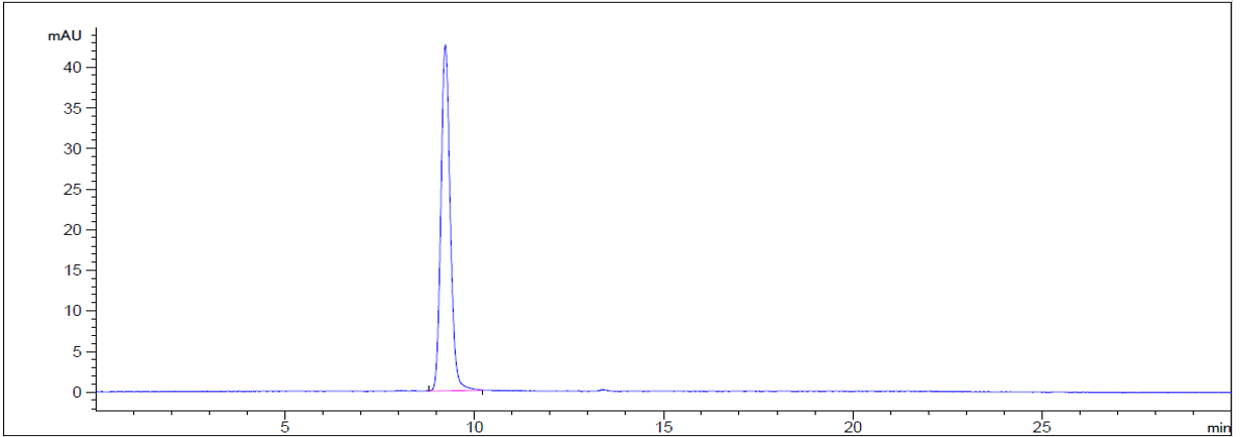
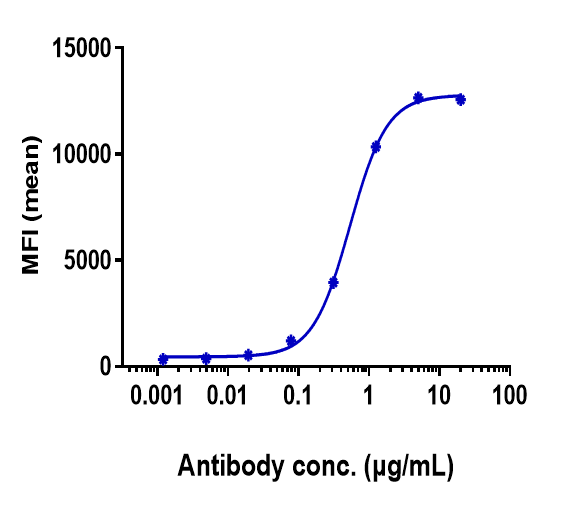
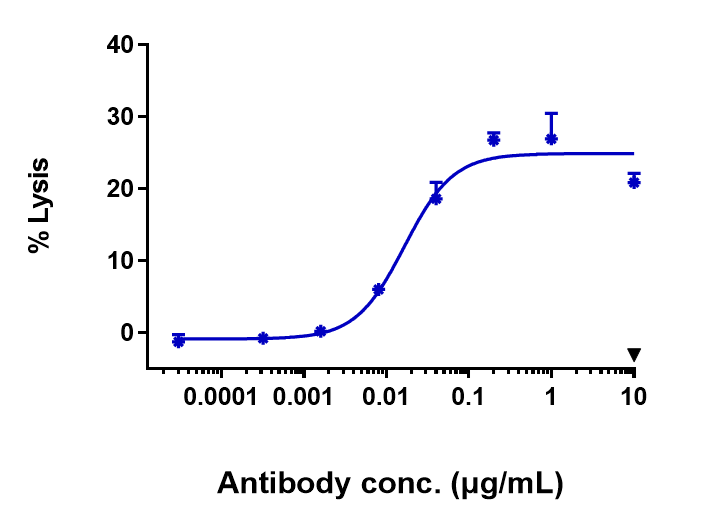
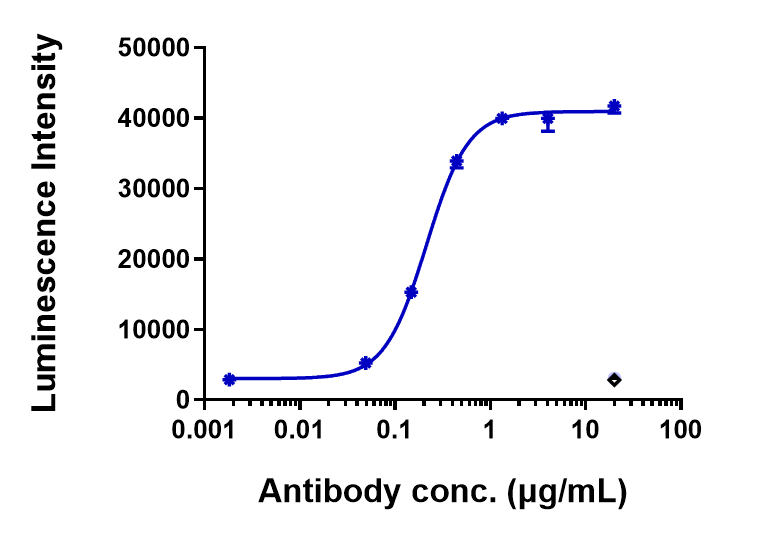
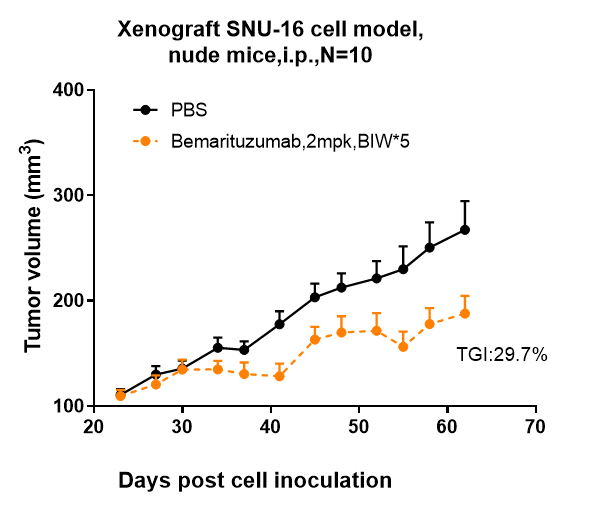
Anti-FGFR2 / CD332 Reference Antibody (bemarituzumab)
Recombinant Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| FC, Kinetics, Animal Model |
|---|---|
| Primary Accession | P21802 |
| Reactivity | Human, Mouse |
| Clonality | Monoclonal |
| Isotype | IgG1 |
| Calculated MW | 144 KDa |
| Target/Specificity | FGFR2 / CD332 |
|---|---|
| Endotoxin | < 0.001EU/ µg,determined by LAL method. |
| Conjugation | Unconjugated |
| Expression system | CHO Cell |
| Format | Purified monoclonal antibody supplied in PBS, pH6.0, without preservative.This antibody is purified through a protein A column. |
| Name | FGFR2 |
|---|---|
| Synonyms | BEK, KGFR, KSAM |
| Function | Tyrosine-protein kinase that acts as a cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation, migration and apoptosis, and in the regulation of embryonic development. Required for normal embryonic patterning, trophoblast function, limb bud development, lung morphogenesis, osteogenesis and skin development. Plays an essential role in the regulation of osteoblast differentiation, proliferation and apoptosis, and is required for normal skeleton development. Promotes cell proliferation in keratinocytes and immature osteoblasts, but promotes apoptosis in differentiated osteoblasts. Phosphorylates PLCG1, FRS2 and PAK4. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. FGFR2 signaling is down-regulated by ubiquitination, internalization and degradation. Mutations that lead to constitutive kinase activation or impair normal FGFR2 maturation, internalization and degradation lead to aberrant signaling. Over-expressed FGFR2 promotes activation of STAT1. |
| Cellular Location | Cell membrane; Single-pass type I membrane protein. Golgi apparatus. Cytoplasmic vesicle. Note=Detected on osteoblast plasma membrane lipid rafts. After ligand binding, the activated receptor is rapidly internalized and degraded [Isoform 3]: Cell membrane; Single-pass type I membrane protein. Note=After ligand binding, the activated receptor is rapidly internalized and degraded [Isoform 13]: Secreted. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.