WWC1 Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC-P, IF, E |
|---|---|
| Primary Accession | Q8IX03 |
| Other Accession | NP_001155133, 242247251 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
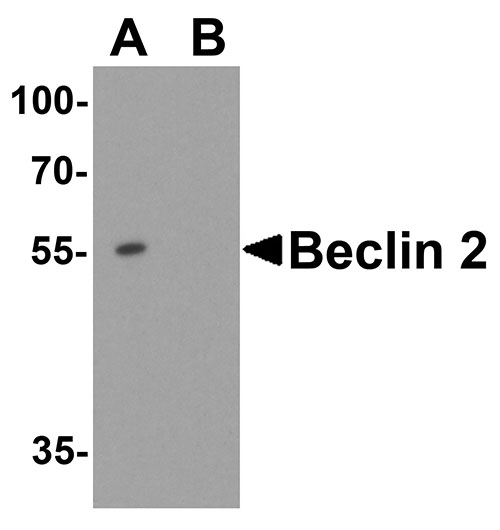
| Calculated MW | Predicted: 123 kDa Observed: 110 kDa |
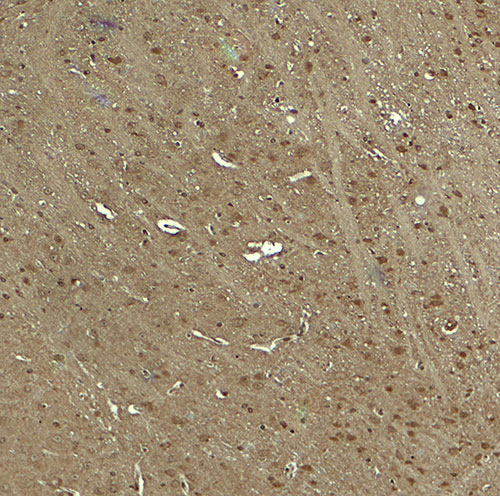
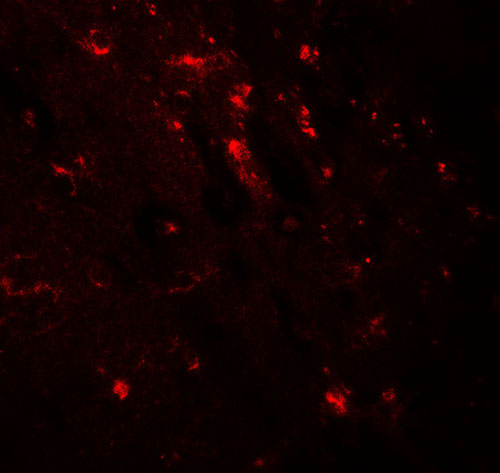
| Application Notes | WWC1 antibody can be used for detection of WWC1 by Western blot at 1 - 2 µg/ml. Antibody can also be used for immunohistochemistry starting at 5 µg/mL. For immunofluorescence start at 20 µg/mL. |
| Gene ID | 23286 |
|---|---|
| Target/Specificity | WWC1; WWC1 antibody is human, mouse and rat reactive. At least three isoforms of WWC1 are known to exist; this antibody will detect all three isoforms. |
| Reconstitution & Storage | WWC1 antibody can be stored at 4℃ for three months and -20℃, stable for up to one year. |
| Precautions | WWC1 Antibody is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | WWC1 (HGNC:29435) |
|---|---|
| Synonyms | KIAA0869, KIBRA |
| Function | Regulator of the Hippo signaling pathway, also known as the Salvador-Warts-Hippo (SWH) pathway (PubMed:24682284). Enhances phosphorylation of LATS1 and YAP1 and negatively regulates cell proliferation and organ growth due to a suppression of the transcriptional activity of YAP1, the major effector of the Hippo pathway (PubMed:24682284). Along with NF2 can synergistically induce the phosphorylation of LATS1 and LATS2 and function in the regulation of Hippo signaling pathway (PubMed:20159598). Acts as a transcriptional coactivator of ESR1 which plays an essential role in DYNLL1-mediated ESR1 transactivation (PubMed:16684779). Regulates collagen-stimulated activation of the ERK/MAPK cascade (PubMed:18190796). Modulates directional migration of podocytes (PubMed:18596123). Plays a role in cognition and memory performance (PubMed:18672031). Plays an important role in regulating AMPA-selective glutamate receptors (AMPARs) trafficking underlying synaptic plasticity and learning (By similarity). |
| Cellular Location | Cytoplasm, perinuclear region. Nucleus. Cell projection, ruffle membrane. Cytoplasm, cytosol. Note=Colocalizes with PRKCZ in the perinuclear region |
| Tissue Location | Expressed in mammary epithelial cells and breast cancer cell lines. Found in the luminal epithelium surrounding the ducts in the normal breast. In the brain, expressed in somatodendritic compartment of neurons in the cortex and hippocampus and in the cerebellum it is found in the Purkinje cells and some granule cells (at protein level). Detected in brain, heart, colon and kidney. In the kidney, expressed in glomerular podocytes, in some tubules and in the collecting duct. |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
The WW and C2 domain containing 1 (WWC1) protein, also known as KIBRA, possesses two WW domains and an internal C2-like domain (1). WWC1 was originally identified as a memory performance-associated protein in humans (2) and has recently been shown to be a novel regulator of the Hippo pathway (3). WWC1 is phosphorylated by the mitotic kinases Aurora-A and –B (4), and in turn activates the Aurora kinases and is required for precise chromosome alignment during mitosis (5).
References
Kremerskothen J, Plaas C, Buther K, et al. Characterization of KIBRA, a novel WW domain-containing protein. Biochem. Biophys. Res. Commun. 2003; 300:862-7.
Papassotiropoulos A, Stephan DA, Huentelman MJ, et al. Common Kibra alleles are associated with human memory performance. Science 2006; 314:475-8.
Genevet A, Wehr MC, Brain R, et al. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 2010; 18:309-16.
Xiao L, Chen Y, Ji M, et al. KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. J. Biol. Chem. 2011; 286:36304-15.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.